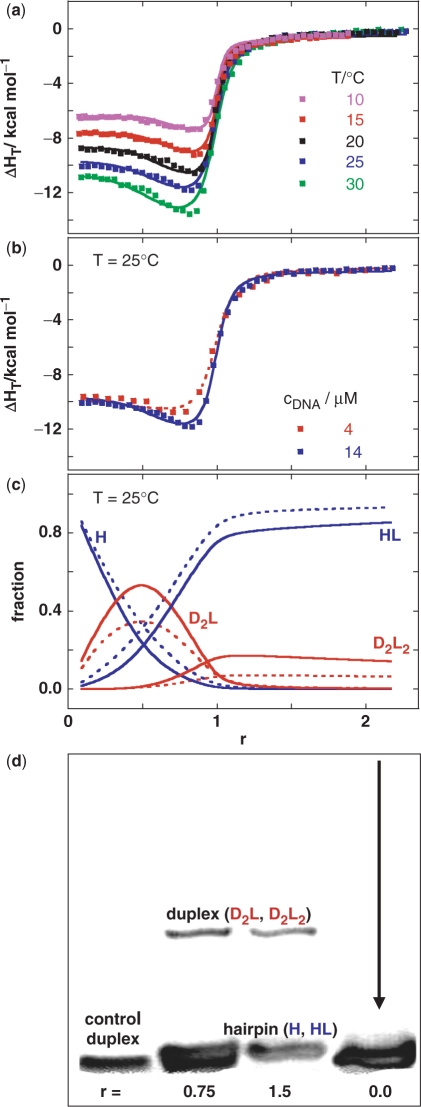
Figure 4.
Model analysis of anomalous heats of ligand–DNA association is supported by the gel electrophoresis experiment. (a) Enthalpies of netropsin association with the model DNA measured by ITC at various netropsin/DNA (single strand) molar ratios (r) and temperatures (cDNA ∼15 μM) and the corresponding best global fit (lines) of the model [Figure 1, Equation (1)]. The best-fit parameters are presented in Supplementary Table 1. (b) ITC curves (symbols) measured at 25°C at cDNA = 4 μM and 14 μM. Dotted line (4 μM) and full line (14 μM) are model functions calculated from the best-fit parameters presented in Supplementary Table 1. (c) Distribution functions of species H, HL, D2L and D2L2 (Figure 1) at cDNA = 4 μM (dotted line) and 14 μM (full line) obtained from the best-fit model thermodynamic parameters (Supplementary Table 1). The fraction of HL is corrected for the small contribution of HL2 species (non-specific binding) and actually represents the sum of the HL and HL2 fractions. The non-specific (low affinity) binding step HL + L ↔ HL2 was added to the model in order to achieve its better agreement with the ITC curves at r > 1. Thermodynamic parameters accompanying the other binding events (Figure 1) are not significantly correlated to parameters required for description of the HL + L ↔ HL2 step (see Supplementary Data). (d) PAGE mobility pattern of model DNA (T = 18°C) mixed with netropsin at different netropsin/DNA molar ratios (r). Control DNA = self-complementary duplex formed from two 5′-CGGAATTCCG-3′ strands should show approximately the same mobility as the hairpin form H (Figure 1).