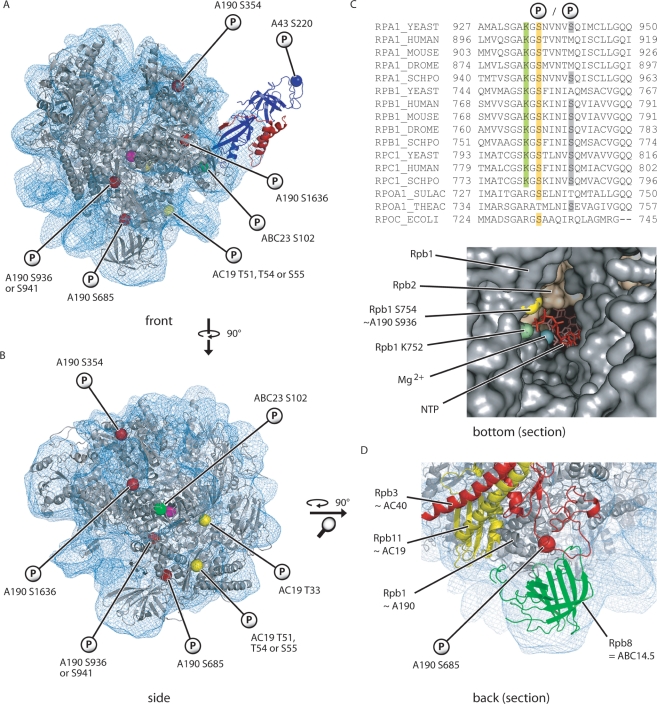
Figure 3.
Three-dimensional localization of the identified Pol I phosphorylation sites. Localization of the phosphorylated amino acids in the Pol I homology model (9). The polymerase core and the A43/A14 heterodimer are shown in gray, blue and red, respectively. The fitted Pol I EM-density is depicted by the blue mesh. The catalytic Mg2+-ion was added for better orientation (magenta sphere in the middle of the structure). (A) Front view. (B) Side view without A43/A14. (C) Sequence alignment of the largest subunits of several eukaryotic RNA polymerases, as well as the homologous subunits from two Archaea species and Escherichia coli. Highlighted amino acids are shown in the structure of the pore/funnel-region below. A section of the bottom view of a Pol II elongation complex (PDB 1Y77) is presented with the downstream DNA on the right hand side. Rpb1 A759 is not located directly on the surface and thus not shown. (D) Localization of A190 S685 on the backside of the polymerase. A magnification of the lower part of the homology model is shown.