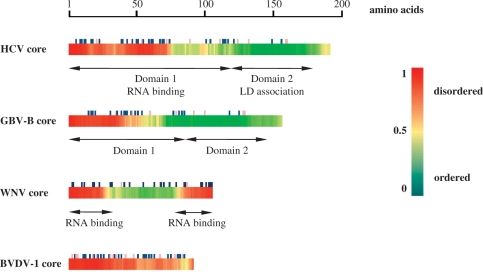
Figure 1.
Disorder prediction in Flaviviridae core proteins. Disordered regions in HCV, GBV-B, WNV and BVDV core proteins (accession numbers D89872; AF179612; AF481864; and AF220247, respectively) were predicted using the DisProt VL3-H predictor (http://www.ist.temple.edu/disprot/predictor.php). An amino acid with a disorder score above 0.5 is considered to be in a disordered environment, while below 0.5 to be ordered. The predicted disorder is illustrated by a colour scale, with highly flexible segments in red and well-folded domains in green. Basic and acidic amino acid residues are indicated by dark blue and mauve symbols, respectively.