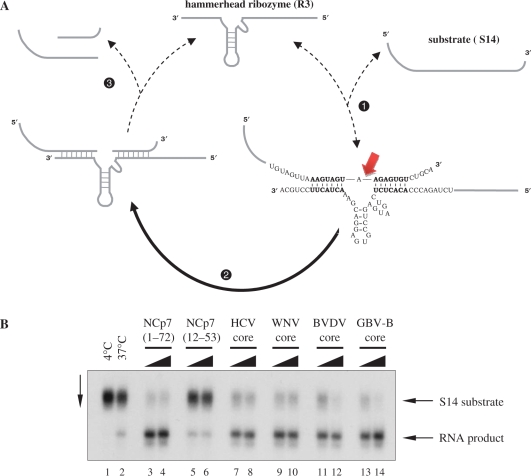
Figure 3.
Enhancement of hammerhead ribozyme cleavage by Flaviviridae core proteins. (A) Schematic representation of the hammerhead ribozyme RNA cleavage reaction. Efficient cleavage of the 32P-labelled RNA substrate (S14) by the R3 hammerhead ribozyme necessitates hybridization of the substrate to the complementary region in the ribozyme sequence (step 1), and release of the cleaved products (step 3), allowing the cyclic reuse of the ribozyme. In the absence of a nucleic acid chaperone, steps 1 and 3 are slow, while both annealing and ribozyme turnover are greatly accelerated by proteins with nucleic acid chaperone activity. S14 RNA substrate cleavage is indicated by the arrow. (B) Hammerhead ribozyme-mediated cleavage is enhanced by Flaviviridae core proteins. R3 ribozyme and S14 substrate RNA were incubated and analysed as described in Materials and Methods section. Lanes 1 and 2: substrate cleavage in the absence of protein at 4°C and 37°C; lanes 3 and 4, 5 and 6, 7 and 8, 9 and 10, 11 and 12 and 13 and 14: substrate cleavage at 37°C in the presence of proteins (as indicated at the top of the figure), at 1:18 and 1:9 protein-to-nucleotide molar ratios (corresponding to 16.6 and 33.3 nM protein concentrations), respectively.