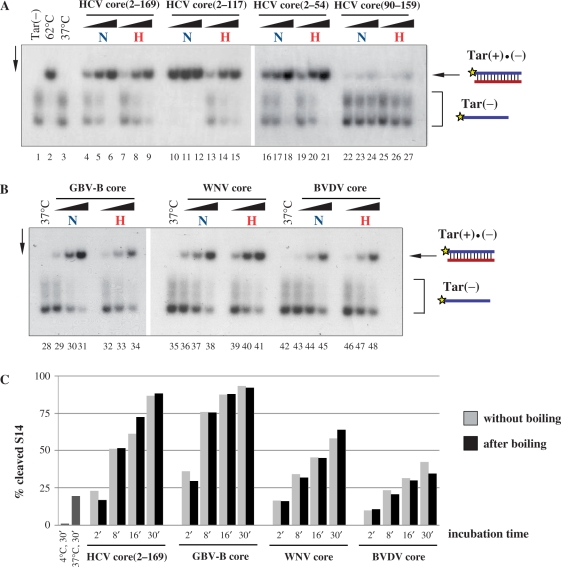
Figure 4.
Heat resistance of Flaviviridae core protein chaperoning activity. (A) and (B) 32P-labelled Tar(−) ODN was incubated together with Tar(+) ODN in the presence of increasing amounts of the proteins, as indicated at the top of the figure. Proteins were either kept on ice (labelled with ‘N’ for native) or boiled (labelled with ‘H’ for heat) for 5 min before mixing with the ODNs. Protein-to-nucleotide molar ratios were 1:40, 1:20 and 1:10 for each protein tested (corresponding to 4, 8 and 16 nM protein concentrations, respectively). Lane 1: labelled Tar(−) ODN alone; lane 2: Tar(−)/Tar(+) complex formed by heat annealing at 62°C, without protein; lanes 3, 28, 35 and 42: Tar(−)/Tar(+) complex formed at 37°C in the absence of protein; lanes 4–6, 10–12, 16–18, 22–24, 29–31, 36–38 and 43–45: complex formation at 37°C in the presence of increasing concentrations of proteins without prior boiling; lanes 7–9, 13–15, 19–21, 25–27, 32–34, 39–41 and 46–48: complex formation at 37°C in the presence of increasing concentrations of proteins after boiling. (C) Kinetics of hammerhead ribozyme cleavage in the presence of Flaviviridae core proteins. R3 ribozyme and 32P-labelled S14 substrate were incubated with core proteins at 1:20 protein-to-nucleotide molar ratio (corresponding to 15 nM protein concentrations) at 37°C. Reactions were stopped at different time points, as indicated in the figure. Proteins were either kept on ice (grey bars) or boiled for 5 min (black bars) before incubation with the RNAs. RNAs were resolved on an 8% denaturing polyacrylamide gel and the percentages of the cleaved S14 substrate were determined by autoradiography and PhosporImager quantification. As a control, R3 and S14 were co-incubated without protein either at 4°C or at 37°C (dark grey bars). Results of a representative experiment are shown.