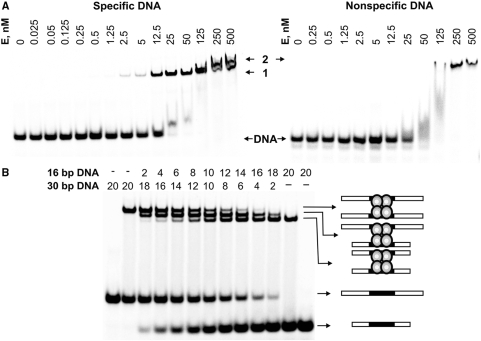
Figure 3.
DNA binding by Cfr42I endonuclease. (A) DNA-binding analysis by gel mobility-shift assay. The reactions contained 1 nM of the 33P-labelled specific oligoduplex 30/30 or the non-specific oligonucleotide NS (see Table 1 for sequence details), and the protein at concentrations (in terms of nM tetramer) as indicated above each lane. After 15 min at room temperature, the samples were subjected to PAGE for 2 h and analysed as described in ‘Materials and Methods’. (B) Analysis of the Cfr42I-DNA complex. Cfr42I (12.5 nM of tetramer) was added to the 30/30 or 16/16 33P-labelled specific DNA duplexes (Table 1) alone or to the duplex mixtures in ratios varying from 9:1 to 1:9 keeping the total DNA concentration fixed at 20 nM. Concentrations of both duplexes in the reactions are indicated above each line. The extreme left and right gel lanes contained no protein. After 15 min at room temperature, the samples were subjected to PAGE for 3 h and analysed as described in ‘Materials and Methods’. The cartoons illustrate protein-DNA complexes that correspond to each band.