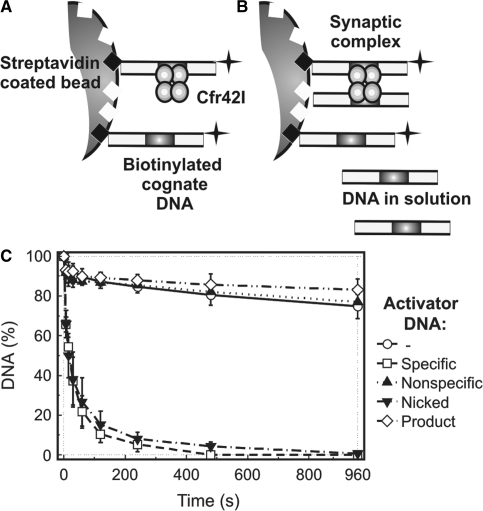
Figure 5.
Immobilized DNA cleavage by Cfr42I. Cartoons in (A) and (B) depict the experimental strategy (36). (A) Oligonucleotide duplex bio-30-30 (Table 1), which carries a biotin at the 5′-end and a 32P label at the 3′ end, is immobilized on the streptavidin-coated magnetic beads at low density. These reaction conditions prevent formation of synaptic complexes between tetrameric REase and two DNA molecules and thus reveal catalytic activity of enzyme bound to a single DNA site. (B) Alternatively, addition of non-biotinylated oligonucleotide duplex (activator DNA) into the reaction mixture enables formation of synaptic complexes between the enzyme, the immobilized DNA and the activator DNA in solution. (C) Experimental results. Upon addition of 25 nM of tetrameric Cfr42I, cleavage of the immobilized hairpin duplex (∼1 nM) was monitored by removing samples at timed intervals (open circles) and analysing them as described in ‘Materials and Methods’. Cleavage of immobilized substrate was also performed in the presence of various non-biotinylated activator DNAs: 50 nM non-specific duplex NS (filled triangles), 50 nM cognate duplex 30/30 (open squares), 50 nM product DNA 16/14p (open diamonds) and 50 nM cognate nicked duplex 30/(14p_16) (inverted filled triangles) (see Table 1 for oligonucleotide sequences). All data points are presented as mean values from ≥3 independent experiments ±1SD.