Figure 3.
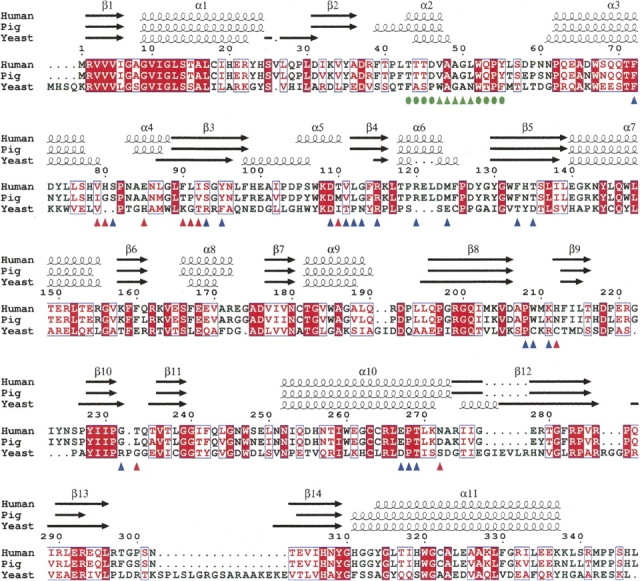
Structure-based sequence alignment of DAO. DAO sequences from human (Swiss-Prot no. P14920), pig (Swiss-Prot no. P00371), and yeast (Swiss-Prot no. P80324) were aligned using ClustalW 1.82 (Thompson et al. 1994) and colored using ESPript 2.2 (Gouet et al. 2003). White in a red box shows strict identity; red in a white box shows similarity within a group; and blue in a white box shows similarity across groups. The hydrophobic stretch is indicated (green triangles) with the residues locally conserved between the human and porcine enzymes (green circles). At the dimer interface of human DAO, 10 residues (red triangles) differ from the porcine enzyme, while 20 residues (blue triangles) are conserved.
