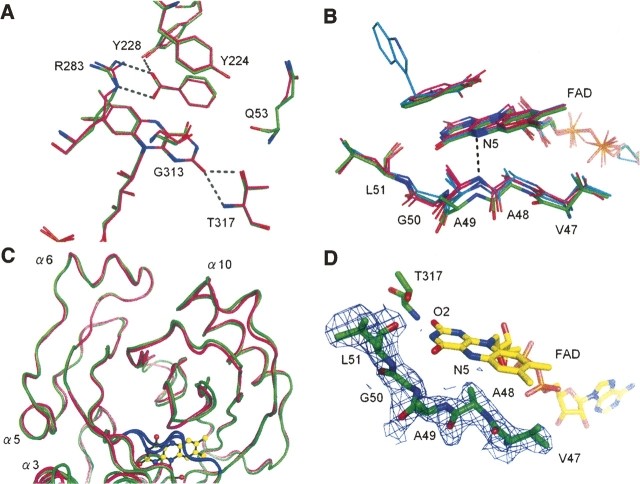
Figure 5.
Context-dependent conformational variability of the hydrophobic stretch. (A) At the re-face of the flavin ring, the active sites are conserved between the human (carbons in green) and porcine (carbons in magenta; PDB code 1VE9) enzymes. H-bonds are shown as dashed lines. (B) Context-dependent conformational variability of the hydrophobic stretch (residues 47–51) at the si-face of the flavin ring. The hydrophobic stretches of human (carbons in green) and porcine (carbons in magenta; PDB codes 1KIF, 1VE9) DAO showed differing conformations, despite strict sequence identity. The purple intermediate of porcine DAO or the structure in complex with imino tryptophan are also superimposed (carbons in cyan). For clarity, only the H-bond between the flavin N5 atom and the Ala49 backbone N atom in the porcine enzyme (PDB code 1KIF) is shown (dashed line). (C) The conformational difference of the hydrophobic stretch (colored in blue) is evident compared with the overall structural similarity between the human (green) and porcine (magenta) DAO structures. (D) The hydrophobic stretch of human DAO is shown with an F o–F c omit map contoured at 1.0 σ.