Abstract
High hydrostatic pressure (HHP)-mediated solubilization and refolding of five inclusion bodies (IBs) produced from bacteria, three Gram-negative binding proteins (GNBP1, GNBP2, and GNBP3) from Drosophila, and two phosphatases from human were investigated in combination of a redox-shuffling agent (2 mM DTT and 6 mM GSSG) and various additives. HHP (200 MPa) combined with the redox-shuffling agent resulted in solubilization yields of ~42%–58% from 1 mg/mL of IBs. Addition of urea (1 and 2 M), 2.5 M glycerol, L-arginine (0.5 M), Tween 20 (0.1 mM), or Triton X-100 (0.5 mM) significantly enhanced the solubilization yield for all proteins. However, urea, glycerol, and nonionic surfactants populated more soluble oligomeric species than monomeric species, whereas arginine dominantly induced functional monomeric species (~70%–100%) to achieve refolding yields of ~55%–78% from IBs (1 mg/mL). Our results suggest that the combination of HHP with arginine is most effective in enhancing the refolding yield by preventing aggregation of partially folded intermediates populated during the refolding. Using the refolded proteins, the binding specificity of GNBP2 and GNBP3 was newly identified the same as with that of GNBP1, and the enzymatic activities of the two phosphatases facilitates their further characterization.
Keywords: high hydrostatic pressure, refolding, inclusion body, solutes, redox-shuffling agent, arginine
Hydrostatic high pressure (HHP) is emerging as a powerful tool for disaggregation and refolding of proteins from insoluble aggregates including amorphous precipitates, amyloid fibrils, and inclusion bodies (IBs) (for review, see Randolph et al. 2002; Kim et al. 2005). For example, HHP processing led to high recovery (>90%) of native protein from aggregates of recombinant human growth hormone, IBs of β-lactamase, and disulfide bond cross-linked aggregates of lysozyme, even at high-protein concentrations (1–8 mg/mL) (St. John et al. 1999, 2001, 2002). HHP disaggregates and refolds proteins from aggregates by disfavoring intermolecular hydrophobic and electrostatic interactions (Randolph et al. 2002; Kim et al. 2005) because hydration of hydrophobic and charged residues reduces system volume (Van Eldik et al. 1989; Silva and Weber 1993). In contrast, hydrogen bonds are not sensitive to HHP due to the negligible volume change associated with breaking of these bonds (Van Eldik et al. 1989; Randolph et al. 2002). To facilitate disruption of hydrogen bonding between protein molecules within aggregates, the temperature can be raised on pressurized samples and/or chaotropes such as guanidine HCl (GdnHCl) and can be included in the protein solution (St. John et al. 1999, 2001, 2002). HHP also cannot break disulfide bonds that sometimes covalently cross-link protein aggregates (Randolph et al. 2002). In these cases, redox-shuffling reagents are included in the pressurized solution to facilitate breaking of intermolecular disulfide bonds and reshuffling of nonnative disulfide bonds into native ones (St. John et al. 1999, 2001, 2002; Randolph et al. 2002).
The effects of numerous solution additives on refolding yields of native protein have been tested with traditional refolding protocols at atmospheric pressure (for review, see De Bernardez Clark et al. 1999; Middelberg 2002). In these approaches, IBs are first dissolved in high concentrations of chaotrope (e.g., 6 M GdnHCl). Then, refolding is fostered by reducing the concentration of chaotrope by dilution and/or dialysis. Often the yield of native protein is low because of (re)aggregations that compete with the refolding (De Bernardez Clark et al. 1999; Middelberg 2002; Morais et al. 2005). To reduce protein aggregations during the refolding, various agents have been tested including low concentrations of chaotropes (GdnHCl and urea), amino acids (arginine), surfactants (Triton X-100, Tween 20, CHAPS), and compatible osmolytes (sucrose, glycerol, sorbitol) (Wetlaufer and Xie 1995; Yasuda et al. 1998; De Bernardez Clark et al. 1999; Middelberg 2002; Ho and Middelberg 2004). Although the mechanisms of action of these compounds are not well understood, empirical screening of solution additives has led to formulations that substantially increase the refolding yield at atmospheric pressure (De Bernardez Clark et al. 1999; Middelberg 2002).
However, there is a paucity of information in the literature on the effects of these different additives on refolding yields obtained with HHP processing of aggregates. Also, published studies on the HHP-mediated refolding have focused mainly on aggregates induced in vitro by stresses such as high temperature and agitation (St. John et al. 1999, 2001, 2002; Foguel et al. 2003; Lefebvre and Robinson 2003). There has been a published report on the refolding of β-lactamase from IBs (St. John et al. 1999) and another describing the refolding of bikunin from soluble oligomers produced in a Chinese hamster ovary cell culture (Seefeldt et al. 2004).
The aim of this study was to determine the effects of solution additives on HHP-mediated solubilization and refolding from IBs of five different eukaryotic proteins produced in Escherichia coli. The model proteins were Gram-negative bacteria binding proteins GNBP 1, GNBP2, and GNBP3 from Drosophila melanogaster (Kim et al. 2000), protein tyrosine phosphatase receptor type S (PTPRS) (Wagner et al. 1996), and dual specificity phosphatase 7 (DUSP7) (Muda et al. 1996) from human. The additives tested were urea, arginine, glycerol, and the surfactants Tween 20 and Triton X-100. Protein yields were assessed by the recovery of total soluble protein, size exclusion chromatography (SEC), and activities of the solubilized proteins.
Results
Descriptions of model proteins
Three Gram-negative binding proteins (GNBPs) from fruit fly and two phosphatases from human, the structural and functional studies of which have been poorly studied due to difficulties in their preparation in soluble forms, were used as model proteins (Table 1). In insects, GNBPs are pattern recognition receptors, which recognize the pattern motifs of invading microbial cell wall components, such as lipopolysaccharide (LPS) from Gram-negative bacteria, peptidoglycan from Gram-positive bacteria, and β-1,3-glucan from yeast and fungi (Royet et al. 2005). After microbial recognition, GNBPs activate innate immune signaling cascades through the Toll pathway for the induction of antimicrobial peptide genes (Kim et al. 2000; Royet et al. 2005). The Drosophila genome encodes three GNBP family proteins (GNBP1, GNBP2, and GNBP3) (Table 1) (Kim et al. 2000; Werner et al. 2000). However, only the binding pattern of GNBP1 expressed in a Drosophila cell has been known (Kim et al. 2000). Here, the extracellular domains of three GNBPs were expressed as IBs and refolded to elucidate their functionality. The sequence alignment shows that GNBP1 shares about 20% and 17% identity with GNBP2 and GNBP3, respectively, and GNBP2 shared about 25% with GNBP3.
Table 1.
Summary of physical and chemical properties of the model proteins
| Proteins (accession code) | Source | MWa (kDa) | pIa | No. of Cys | Hydropathicitya | SVCb (mL mol/g2) |
| GNBP1 (NP_524142) | D. melanogaster | 52.1 | 5.26 | 6 | −0.463 | 0.87 |
| GNBP2 (NP_524141) | D. melanogaster | 50.0 | 5.81 | 8 | −0.38 | 0.49 |
| GNBP3 (NP_523986) | D. melanogaster | 51.0 | 5.88 | 7 | −0.635 | 1.80 |
| PTPRS (NP_002841) | Human | 64.9 | 6.41 | 11 | −0.456 | 0.45 |
| DUSP7 (NP_001938) | Human | 35.1 | 5.56 | 9 | −0.291 | 0.47 |
a The ProtParam tool of the ExPASy proteomics server of the Swiss Institute of Bioinformatics (http://kr.expasy.org/tools/protparam.html) was used for the determination of the molecular weight (MW), theoretical pI, and hydropathicity based on the scale developed by Kyte and Doolittle (1982).
b Second virial coefficients (SVCs) was estimated by the following formula (Ho and Middelberg 2004)): SVC (mL mol/g2) = −0.0299 (MW) −5.233H, where MW is molecular weight of proteins in kDa and H is hydropathicity.
Protein tyrosine phosphatase receptor type S (PTPRS), belonging to a family of the protein tyrosine phosphatase, are signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation (Wagner et al. 1996). The intracellular phosphatase domain of PTPRS was expressed and studied here. Dual specificity phosphatase 7 (DUSP7), a member of the mitogen-activated protein kinase family, is an intracellular phosphatase with a dual specificity of removing the phosphoryl group from both Tyr and Thr residues (Muda et al. 1996). The sequence analysis reveals that PTPRS shares about 12% with DUSP7.
Effects of additives on the solubilization and refolding of proteins from IBs
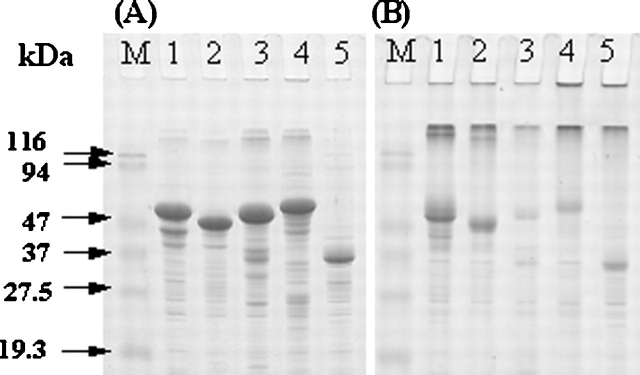
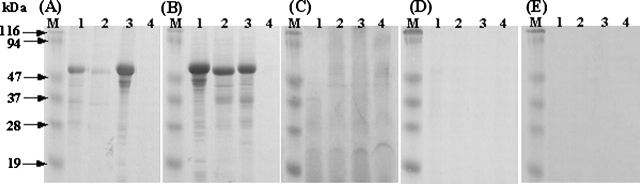
Each of the model proteins contains more than six Cys residues (Table 1). When analyzed by SDS-PAGE, the purified IBs showed a main band at the expected position for each protein under the reducing condition (Fig. 1A). In contrast, for all of the proteins, large aggregates accumulated at the interface of stacking and separating gels with minor fractions of monomeric species under the nonreducing condition (Fig. 1B). Densitometric analyses of SDS–polyacrylamide gels stained with Coomassie blue estimated ~32%, 26%, 6%, 11%, and 39% of monomeric species for GNBP1, GNBP2, GNBP3, PTPRS, and DUSP7, respectively, in the nonreducing condition. Thus, as has been observed with other proteins (De Bernardez Clark et al. 1999; Middelberg 2002), the IBs contain nonnative intermolecular disulfide bond cross-linked protein aggregates, even though they formed in the reducing cytosolic environment of bacteria.
Figure 1.
Reducing (A) and nonreducing (B) SDS-PAGE analysis of the purified IBs: GNBP1 (lane 1), GNBP2 (lane 2), GNBP3 (lane 3), PTPRS (lane 4), and DUSP7 (lane 5). About 5 μg of each IB was analyzed on 12% SDS-PAGE under reducing and nonreducing conditions. The gels were stained with Coomassie blue. The molecular mass markers are indicated in kilodaltons.
To allow nonnative disulfide bonds to be reshuffled into native ones during the high hydrostatic pressure (HHP)-assisted dissolution and refolding of protein from IBs, the refolding buffer included the redox-shuffling agent (2 mM DTT and 6 mM GSSG) at pH 8.0 (St. John et al. 2002). Instead of GSH, DTT was used to reduce the total glutathione concentration (De Bernardez Clark et al. 1998). An alkaline pH above at least pH 7.5 is required to promote thiolate anion formation for reshuffling of disulfide bonds (De Bernardez Clark et al. 1999; Middelberg 2002; Kim et al. 2005). Other previous studies have shown that the refolding efficiency generally is optimal at the conditions of pressure and temperature that thermodynamically favor the protein’s native structure, i.e., inside (the Gibbs free-energy change of denaturation [ΔG] >0) of the elliptic pressure-temperature phase diagram (Hawley 1971; Randolph et al. 2002). Native-state proteins are usually stable up to 300 MPa at 25°C (Gross and Jaenicke 1994; Randolph et al. 2002; Winter and Dzwolak 2005). Thus, refolding experiments started with incubations of IBs (1 mg/mL) under 200 MPa at 25°C for 24 h in the refolding buffer. Further, the effects of various additives, such as urea (1 and 2 M), arginine (0.5 M), and glycerol (2.5 M), on the solubilization and refolding from IBs were tested by using their typical ranges of concentrations as used in the literatures, i.e., ≤2 M urea, 0.4–0.8 M arginine, and 0.4–3 M glycerol (De Bernardez Clark et al. 1999; Middelberg 2002; Ho and Middelberg 2004).
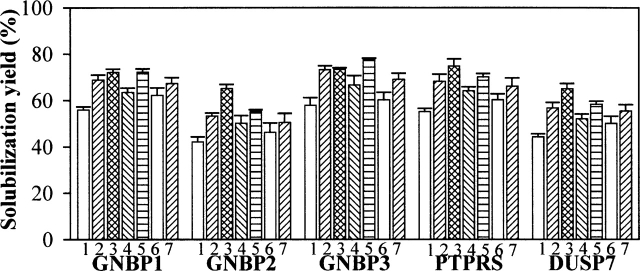
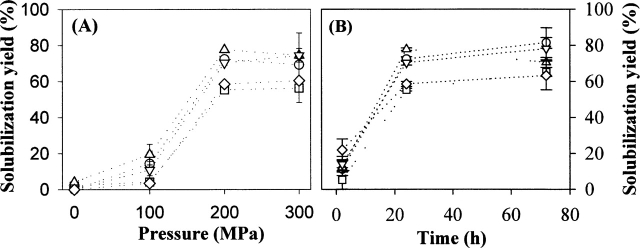
After pressure release, samples were centrifuged to remove insoluble aggregates, and the supernatants were used for the total protein assay to determine “solubilization yield.” Control samples incubated at atmospheric pressure (0.1 MPa) under the above buffer conditions exhibited minimal or no solubilization (0%–3%) for all proteins (data not shown). However, the HHP treatment in the refolding buffer led to solubilization yields ranging from 42% (GNBP2) to 58% (GNBP3) (Fig. 2). Solubilization yields by HHP in the TBSE buffer without the redox-shuffling agent ranged from ~5% to 20% (data not shown). The presence of additives increased solubilization yields to ~55%–78% for all of the proteins (Fig. 2). Of the solution conditions tested, 2 M urea and 0.5 M arginine provided the greatest enhancement of solubilization (Fig. 2).
Figure 2.
HHP mediated-solubilization yield of IBs (1 mg/mL) incubated under 200 MPa at 25°C for 24 h in the refolding buffer (50 mM Tris-Cl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, 6 mM GSSG, 0.05% sodium azide) containing the various additives. The numbers below each column represent the buffer formulation as follows: (1) buffer alone; (2) 1 M urea; (3) 2 M urea; (4) 2.5 M glycerol; (5) 0.5 M arginine; (6) 0.1 mM Tween 20; (7) 0.5 mM Triton X-100. After decompression, samples were centrifuged (12,000g for 10 min) to remove insoluble aggregates and the supernatants were used for the total protein assay as described in the Materials and Methods in detail. The error bars represent the standard deviation for triplicate incubated samples.
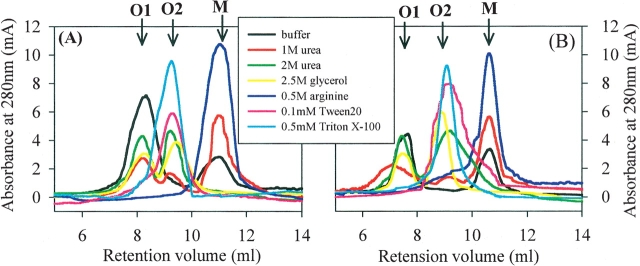
SEC analyses showed that the recovered soluble proteins from IBs (1 mg/mL) were composed of multiple assembly states from monomeric species to oligomeric species (dimeric and/or trimeric species and soluble aggregates), the relative distribution of which varied significantly depending on the additives (Fig. 3). The soluble proteins from the refolding buffer eluted as <30% monomer and >70% soluble aggregates (more than trimeric species), which was eluted in the void volume and designated as “O1 species,” for all of the proteins (Fig. 3; data not shown). The presence of 1 M urea populated more monomer (~55%–70%) and new oligomeric species (~5%–15%), which was estimated to be dimeric and/or trimeric species and designated as “O2 species”, with concomitant decreases of O1 species (~21%–38%), compared with those from the refolding buffer only (Fig. 3). For 2 M urea and 2.5 M glycerol, the fractions of oligomeric O1 and O2 species in the solubilized proteins were ~26%–44% and ~54%–68%, respectively, which were much higher than monomer (~0%–10%) for all proteins. Very interestingly, however, the soluble proteins obtained in the presence of 0.5 M arginine eluted as >95% monomer and <5% O2 species for GNBP1 and GNBP2 and PTPRS, and dominantly monomer (>70%) and O2 species (<30%) for GNBP3 and DUSP7 (Fig. 3; data not shown).
Figure 3.
Representative size exclusion chromatogram for the recovered soluble GNBP1 (A) and PTPRS (B) from IBs (1 mg/mL) by HHP (200 MPa, 25°C, 24 h) in the refolding buffer containing the additives, as described in the legend for Figure 2 and also indicated in the legend box by different colors. The arrows indicate the elution position for monomeric species as M, for dimeric and/or trimeric species as O2, and for soluble aggregates (more than tetramer) as O1.
To test whether the effects of arginine were exerted by its ionic strength effects, samples were refolded by HHP under the conditions described in the refolding buffer containing various NaCl concentrations (150, 300, or 500 mM). The solubilization yields and SEC elution profiles of the recovered soluble proteins were similar to those obtained in refolding buffer alone (data not shown). This result suggests that the effect of arginine was derived from property other than ionic strength. In a previous study of dilution refolding from denatured states, 0.5 M arginine was also more effective in generating active monomeric species of carbonic anhydrase, compared with 0.5 M NaCl and 0.5 M GdnHCl (Baynes et al. 2005).
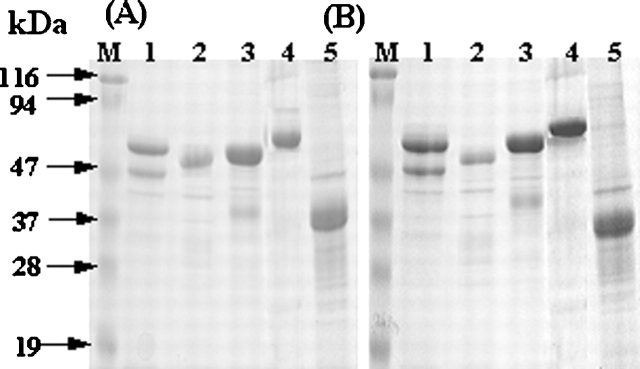
When analyzed by reducing and nonreducing SDS-PAGE, the soluble proteins from the various refolding conditions migrated at their corresponding monomeric position with comparable band intensities in both conditions (Fig. 4; data not shown), suggesting that the soluble oligomeric O1 and O2 species did not contain intermolecular disulfide bonds. Attempts to dissociate oligomers by repressurization (200 MPa, 25°C, 24 h) of the soluble proteins in the buffer with the various additives did not change significantly their oligomeric states (data not shown), suggesting that the species are not pressure sensitive. To test whether nonionic surfactants could induce dissociation of the soluble oligomers, surfactants (0.1 mM Tween 20 and 0.5 mM Triton X-100) were used in the refolding buffer. The surfactants slightly increased the solubilization yields for all proteins (Fig. 2), but populated dominantly dimeric and/or trimeric O2 species (>90%) by eliminating the soluble large aggregates of O1 species (<5%) and monomer (~0%) for all proteins (Fig. 3; data not shown).
Figure 4.
Representative reducing (A) and nonreducing (B) SDS-PAGE analysis of the recovered soluble proteins from IBs (1 mg/mL) by HHP (200 MPa, 25°C, 24h) in the refolding buffer. GNBP1 (lane 1); GNBP2 (lane 2), GNBP3 (lane 3), PTPRS (lane 4), and DUSP7 (lane 5). The gels were stained with Coomassie blue.
Functional assays
The recovered soluble proteins were extensively dialyzed against the TBSE buffer and then subjected to their functional assays. For GNBPs, only the functionality of GNBP1 expressed in a Drosophila cell has been known, which specifically recognizes and binds to both β-1,3-glucan and LPS, but not to peptidoglycan, cellulose, or chitin (Kim et al. 2000). The recovered soluble GNBPs (~10–15 μM) from the refolding buffer containing 0.5 M L-arginine were used for the binding assay, with BSA (20 μM) as a negative control. Figure 5 shows that all GNBPs bound to only β-1,3-glucan and LPS, but not to the other substrates, demonstrating that the pattern of substrate recognition for GNBP2 and GNBP3 is the same with that of GNBP1 (Kim et al. 2000). The specific binding activities of the soluble GNBPs also indicate that the monomers are the functional, native species with proper conformations. The binding assay was not reproducible with the soluble proteins recovered after pressure treatment in the other refolding formulations.
Figure 5.
Functional binding assay of the recovered soluble GNBPs from their respective IBs (1 mg/mL) by HHP (200 MPa, 25°C, 24 h) in the refolding buffer containing 0.5 M arginine. The binding assay was performed as described in the Materials and Methods in detail using the various microbial cell wall components, β-1,3-glucan (A), LPS (B), chitin (C), cellulose (D), and peptidoglycan (E). GNBP1 (lane 1); GNBP2 (lane 2); GNBP3 (lane 3); and BSA as a negative control (lane 4).
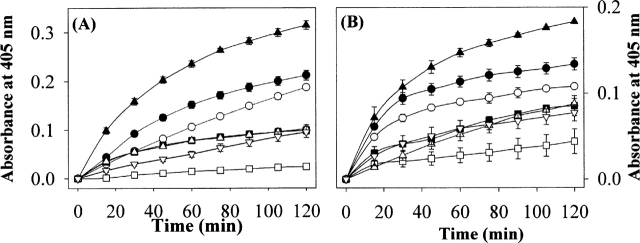
The enzymatic activities of the pressure solubilized phosphatases (~5 μM), PTPRS and DUSP7 varied dramatically depending on the refolding buffer conditions, with activity in the order of 0.5M arginine > 1 M urea> buffer alone > glycerol ≈ surfactants > 2 M urea (Fig. 6). The magnitude of the observed phosphatase activities across the buffer conditions were proportional to the contents of monomeric species in each refolding buffer analyzed by SEC (Fig. 3), suggesting that the monomeric species are functional with correct conformations.
Figure 6.
The phosphatase activity assay of the recovered soluble PTPRS (A) and DUSP7 (B) from IBs (1 mg/mL) by HHP (200 MPa, 25°C, 24 h) in the refolding buffer containing the additives. The enzymatic activity was monitored by the p-NPP hydrolysis assay as described in detail in the Materials and Methods section. The symbols represent the following buffer conditions: (○) buffer; (•) 1 M urea; (□) 2 M urea; (▪) 2.5 M glycerol; (▴) 0.5 M arginine; (▵) 0.1 mM Tween 20; (▿) 0.5 mM Triton X-100. The error bars represent the standard deviation for triplicate incubated samples.
Effects of pressure and duration of pressurization
Previous studies have shown that moderate HHP of 100–300 MPa is generally effective for dissociating protein oligomers and aggregates, while relatively higher HHP (>300 MPa) is usually required for the denaturation of native monomeric proteins (Hawley 1971; Silva and Weber 1993; Randolph et al. 2002; Winter and Dzwolak 2005). To access the effect of pressure on the protein refolding, IBs (1 mg/mL) were pressurized to 100, 200, or 300 MPa at 25°C for 24 h in the refolding buffer with or without 0.5 M arginine. The maximal recovery of soluble proteins was achieved at 200 MPa with similar yields at 300 MPa and significantly lower yields at 100 MPa for all proteins. The presence of 0.5 M arginine slightly improved the solubilization yield at each HHP (Fig. 7A). The higher pressure of 300 MPa did not significantly change the relative ratio of monomer to soluble oligomeric species in the soluble proteins, compared with those at 200 MPa (data not shown).
Figure 7.
Effects of pressure (A) and duration of pressurization (B) on the solubilization yield of IBs in the refolding buffer containing 0.5 M arginine. In A, IBs (1 mg/mL) were incubated under the designated HHP at 25°C for 24 h. In B, IBs (1 mg/mL) were incubated under 200 MPa at 25°C for 2, 24, and 72 h. The symbols represent each protein as follows: GNBP1 (○); GNBP2 (□); GNBP3 (▵), PTPRS (▿), and DUSP7 (⋄). The error bars represent the standard deviation for triplicate incubated samples.
To determine the effects of duration of HHP on the degree of solubilization yield, IBs (1 mg/mL) in the refolding buffer with 0.5 M arginine were pressurized at 200 MPa at 25°C for 2, 24, and 72 h. Figure 7B shows that the solubilization yields in 24 h exhibited comparable values in 72 h for all proteins, suggesting that the solubilization kinetics of proteins reach steady-state conditions at ~24 h, as was found previously with recombinant human growth hormone (St. John et al. 1999) and lysozyme (St. John et al. 2002).
Relationship between atmospheric urea solubility and solubilization yield by HHP
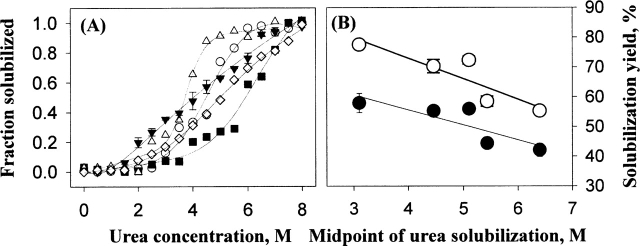
As stated earlier, HHP dissolves protein aggregates by disrupting mainly hydrophobic and electrostatic interactions, but is neutral to hydrogen bonds (Silva and Weber 1993; Randolph et al. 2002; Kim et al. 2005). On the other hand, urea solubilizes protein aggregates by breaking both hydrogen bonding and hydrophobic interactions within the aggregates (De Bernardez Clark et al. 1999; Middelberg 2002). To elucidate any relationships between atmospheric urea solubility of IBs and the HHP-mediated solubilization yields, the atmospheric urea solubility of IBs (0.5 mg/mL) were determined as a function of urea concentration (0–8 M). The calculated midpoint urea concentration showed an inverse relationship (r2 ≈ 0.75) with the HHP-mediated solubilization yields (Fig. 8).
Figure 8.
Atmospheric urea solubility of IBs (A) and the plotting of the midpoint of urea solubilization versus the solubilization yields of IBs (1 mg/mL) by HHP (200 MPa, 25°C, 24 h) (B). In A, the symbols represent each protein as follows: GNBP1 (○); GNBP2 (▪); GNBP3 (▵), PTPRS (▾), and DUSP7 (⋄). In B, closed (•) and open (○) symbols represent the solubilization yields of IBs incubated in the refolding buffer alone and containing 0.5 M arginine, respectively. The error bars represent the standard deviation for triplicate incubated samples.
Discussion
HHP efficiently disaggregated and refolded five different proteins from IBs produced from bacteria (Figs. 2–6). All of the solution additives tested greatly increased solubilization yields for all of the proteins (Fig. 2). However, the recovery of the functional monomeric species was dramatically dependent on the specific additive (Figs. 3–6). A critical question is whether the soluble aggregates are formed under HHP from larger aggregates and/or during depressurization from monomers produced by HHP. IBs are usually formed by the aggregation of partially folded intermediates of proteins (De Bernardez Clark et al. 1999; Middelberg 2002). The initial events of the HHP-mediated refolding might be the release of the aggregation-prone, partially folded intermediates from IBs. The HHP refolding conditions used in the current study (200 MPa, 25°C) most likely favor thermodynamically the native over the denatured state, which should facilitate the metastable intermediates proceeding to fold into the native states (Randolph et al. 2002; Kim et al. 2005). However, under some solution conditions, partially folded intermediates, which have more perturbed and solvated structures, and thus smaller volume than the native state, could be populated under HHP, but do not aggregate until pressure is released (Gorovits and Horowitz 1998).
Thus, it is most likely that the observed soluble oligomeric species are formed by reaggregation of the partially folded intermediates during and/or depressurization, rather than under HHP. This argument is supported by the observations that the instant release of HHP significantly decreased the solubilization yields (≤32%) for all proteins (data not shown) and longer pressurization did not affect the solubilization yields (Fig. 7B). Previous studies have also shown that HHP-induced nonnative species of transthyretin (Ferrao-Gonzales et al. 2000) and glutathione reductase (Morais et al. 2005) rapidly formed protein aggregates upon depressurization. Both the solubility and stability of partially folded intermediates can be modulated by changing solution conditions using additives (De Bernardez Clark et al. 1999; Kim et al. 2001; Middelberg 2002). In this respect, a major effect of the additives during HHP treatments is to favor refolding of partially unfolded intermediates over aggregation of these species.
Arginine (0.5 M) combined with HHP (200 MPa) was most effective for obtaining functional monomeric protein molecules from IBs. Arginine has minimal effect on the thermodynamic stability of the native state (Shiraki et al. 2002; Arakawa and Tsumoto 2003), yet it has been shown to enhance greatly the refolding of several proteins from the unfolded states by suppression of aggregation (Arora and Khanna 1996; Armstrong et al. 1999; Baynes et al. 2005; Reddy et al. 2005). The exact mechanism of how arginine functions as a suppressor of protein aggregation is not fully elucidated, but it has been proposed that arginine binds to and enhances the stability of partially folded or denatured proteins by inhibiting nonnative hydrophobic interactions like chaperones, thereby leading to a decrease in their aggregations (Shiraki et al. 2002; Arakawa and Tsumoto 2003; Ishibashi et al. 2005; Reddy et al. 2005). Another theory has proposed that arginine sterically excluded from the interfaces between protein–protein encounter complexes to increase the energetic barrier for protein–protein association, eventually leading to suppression of aggregations between monomeric species (Baynes and Trout 2004; Baynes et al. 2005). The mechanisms proposed above are likely operative during HHP treatment.
Glycerol, nondenaturing low concentrations of urea (≤2M), and nonionic surfactants have also been identified as useful refolding agents for many proteins (Wetlaufer and Xie 1995; De Bernardez Clark et al. 1999; Middelberg 2002; Ho and Middelberg 2004). Previous studies have shown that the combination of HHP (200 MPa) with GdnHCl (0.8 M) and glycerol (2 M) exhibited significant synergistic effects on both solubilization and recovery of active monomeric species of lysozyme (St. John et al. 2002) and rhodanese (Gorovits and Horowitz 1998), respectively. However, our results showed that urea (1–2 M), glycerol (2.5 M), and nonionic surfactants (0.1 mM Tween 20 and 0.5 mM Triton X-100) populated more soluble oligomeric species than monomeric species for all proteins tested under HHP (Fig. 3). Further, these additives induced the formation of new oligomeric O2 species, estimated to be dimeric and/or trimeric species, compared with the results obtained with buffer alone (Fig. 3). The mechanism resulting in this phenomenon is not clear at this point. It seems that, even in the nondenaturing concentrations, urea combined with HHP destabilized proteins to populate aggregation-prone, partially folded intermediates, which failed to proceed to correctly refold under HHP, but instead aggregated upon depressurization. This aspect is supported by the observation that 1 M urea populated more functional monomeric species than 2 M urea, which dominantly generated inactive soluble oligomers (Figs. 3, 6).
Glycerol thermodynamically stabilizes the native state of proteins by preferential exclusion (Timasheff 1998). However, the preferential exclusion can also enhance protein assembly, such as the case of enhanced polymerization of tubulin into microtubules in the presence of glycerol (Sackett 1997). Likewise, it seems that glycerol promoted the oligomerization of partially folded intermediates populated under HHP.
Nonionic surfactants dominantly induced the O2 species estimated to be dimer and/or trimer (Fig. 3). Probably, the surfactants preferentially interact with the oligomers to increase their solubilities, like the case of lysozyme during refolding by various surfactants including Tween 20 (Yasuda et al. 1998). In our previous works, the addition of Tween 20 also caused a greater accumulation of soluble aggregates during reconstitution of lyophilized anti-L-selectin (Jones et al. 2001) and agitation of recombinant human factor XIII (Kreilgaard et al. 1998) compared with the buffer control.
Previously, second virial coefficients (SVC) measured experimentally by static light scattering and estimated by protein sequence for eight IBs exhibited an inverse relationship with the propensity to form protein aggregates during refolding (Ho and Middelberg 2004). When the physicochemical, structural properties of the model proteins (Table 1), such as size, hydropathicity, SVC calculated, and predicted secondary structural contents were plotted against the solubilization yields by HHP, no significant relationships were observed. Instead, the atmospheric urea solubility of IBs were inversely correlated with the HHP-mediated refolding efficiency (Fig. 8), suggesting that intermolecular hydrogen bonding is a main thermodynamic energy barrier in HHP-mediated solubilization of the proteins from IBs (St. John et al. 2001). Arginine could also be effective at disrupting hydrogen bonds under pressure, and thus facilitate the solubilization of aggregates under pressure as well as foster refolding of partially unfolded intermediates over reaggregation during depressurization.
Materials and methods
Materials
L-arginine, urea, glycerol, dithiothreitol (DTT), oxidized glutathione (GSSG), Tween 20, and Triton X-100 were purchased from Sigma-Aldrich Co. Restriction enzymes were purchased from New England Biolabs. All other chemicals were commercially available reagent grade.
Protein expression and purification of IBs
The bacterial expression plasmids encoding the extracellular domain (ECD) of GNBPs were provided by Dr. B.H. Oh (Pohang University of Science and Technology). Briefly, residues 24–492 of GNBP1-ECD and residues 25–461 of GNBP2-ECD were cloned on pProEx-HTb (Invitrogen) using BamHI/XhoI sites, and residues 31–483 of GNBP3-ECD was subcloned into pProExHTc (Invitrogen) using EcoRI/XhoI sites. For the bacterial expression of the intracellular phosphatase domain of PTPRS and DUSP7, residues 1386–1948 of PTPRS and residues 1–130 of DUSP7 were subcloned onto pET28a (Novagen) using NdeI/BamHI sites, which were provided by Dr. J.S. Kim (Korea Research Institute of Bioscience and Biotechnology). E. coli strain BL21(DE3) (Novagen) was used as an expression host. Cells were grown at 37°C to an OD600 of ~0.8 in 100 mL of Luria-Bertani medium containing 100 μg/mL amplicillin, and protein expression was induced by the addition of 1 mM isopropyl-β-D-1-thiogalactoside. After a 6-h induction at 37°C, cells were harvested by centrifugation at 12,000g for 10 min at 4°C and resuspended in a 10-mL lysis buffer (50 mM Tris-Cl at pH 8.0, 5 mM EDTA, 100 mM NaCl, 1 mM PMSF). IBs were purified by following the procedure described previously (Bowden et al. 1991; De Bernardez Clark et al. 1999). The purified IBs were washed three times with double distilled water, and stored at −80°C until used. The contents of the expressed protein in purified IBs were more than 70% based on SDS-PAGE analyses (Fig. 1). After gel staining with Coomassie blue, the intensity of bands corresponding to monomeric proteins in the reducing and nonreducing SDS-PAGE was compared with estimate fractions of monomeric species in the nonreducing condition using an image analysis system (Model GS-700, Bio-Rad).
Formulation of refolding buffers and sample pressurization
IBs were resuspended in a TBSE buffer (50 mM Tris-Cl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.05% sodium azide), containing a redox-shuffling mixture (2 mM DTT and 6 mM GSSG), designated as a “refolding buffer”. The molar ratio of oxidized to reduced glutathione (GSSG:GSG) was 1:1 because the mixture of 2 mM DTT and 6 mM GSSG rapidly converts to 4 mM GSH and 4 mM GSSG, with a byproduct of 2 mM oxidized DTT (St. John et al. 2002). For testing the effects of additives, urea (1 and 2 M), L-arginine (0.5 M), glycerol (2.5 M), Tween 20 (0.1 mM), or Triton X-100 (0.5 mM) were added to the refolding buffer. The critical micelle concentrations (CMCs) of Tween 20 (Sigma T8787) and Triton X-100 (Sigma P5927) provided by the supplier (Sigma) were 0.06 and 0.32 mM in water, respectively.
Samples (~300 μL) were placed in sterile, disposable 1-mL syringes (one end heat sealed and the other sealed with the rubber plunger) and placed in a custom-made high-pressure vessel (St. John et al. 2001; Kim et al. 2005). The vessel was sealed and pressurized with a high-pressure crank generator (rated up to 700 MPa) from High Pressure Equipment Co., using water as a pressure transmitting fluid (St. John et al. 2001; Kim et al. 2005). Samples were slowly pressurized (~10 min) to the desired pressure and held there for 24 h. Unless otherwise specified, samples were depressurized at ~10 MPa per 10 min. All pressure experiments were performed at room temperature (~25°C).
Total protein assay and SEC
After the pressure was released, each sample was removed from the syringe, placed into a microcentrifuge tube, and centrifuged (12,000g for 10 min) to remove insoluble aggregates. The supernatants were assayed for total protein content using a Bradford dye binding assay (Bio-Rad), which according to the manufacturer’s instructions is compatible with the various additives used. To determine the total protein contents of the IB samples, the IBs were solubilized by incubation overnight (25°C) in the refolding buffer containing 9 M urea. Then, the samples were serially diluted with the TBSE buffer to urea concentrations below 6 M, which is compatible with the total protein assay.
The soluble proteins—before or after dialysis against the TBSE buffer using Slide-A-lyzer dialysis cassette (Pierce)—were analyzed by SEC and reducing and nonreducing SDS-PAGE. SEC, which was used to quantify monomer and oligomer levels, was performed on a Pharmacia AKTA-FPLC system using a TSK-GEL G3000SWXL size exclusion column (Tosohaas), with a mobile phase (50 mM sodium phosphate at pH 7.5, 150 mM NaCl) at a flow rate of 0.7 mL/min. The injection volume of the sample was 50 μL, and the sample elution was monitored by absorbance at 280 nm. Peak areas were computed using the vendor-supplied software.
Functional assays of solubilized proteins
After depressurization and subsequent centrifugation, the soluble proteins were dialyzed against the TBSE buffer and then assayed for functional activity. The binding assays of GNBPs were carried out essentially as described previously (Kim et al. 2000) using the substrates, cellulose (β-1,4-glucan) (Sigma), chitin (β-1,4-N-acetyl-D-glucosamine) (Sigma), peptidoglycan (β-1,4-glycosidic linkage between N-acetylmuramic acid and N-acetylglucosamine) (Fluka), lipopolysaccharide (Sigma), and curdlan (β-1,3-glucan) (Sigma). Briefly, 100 μg of each insoluble substrate was added to 500 μL of the solubilized GNBPs (~10–15 μM), and the mixture was incubated at 25°C for 1 h with mild agitations. Then, samples were centrifuged (12,000g for 5 min) and the pellet was washed twice with 500 mL of TBSE buffer containing 0.02% (v/v) Triton X-100. The proteins bound on the substrates were desorbed by adding SDS-PAGE sample buffer and analyzed by reducing SDS-PAGE (Kim et al. 2000). As a negative control, BSA (20 μM) was used.
The phosphatase activity of PTPRS and DUSP7 was assayed using p-nitrophenyl phosphate (p-NPP) (Sigma) as a substrate (Montalibet et al. 2005). The proteins (~5 μM) were incubated at 37°C with 5 mM p-NPP in 25 mM HEPES (pH 7.2) containing 50 mM NaCl, 5 mM DTT, and 2.5 mM EDTA. The time course of formation of p-NP was monitored every 3 min for 120 min by absorption increase at 405 nm against an enzyme-free blank in a VersaMax microplate reader (Molecular Devices).
Atmospheric urea solubility of IBs
About 0.5 mg/mL of IBs were equilibrated overnight at 25°C in the TBSE buffer containing urea concentrations from 0 to 8 M. After centrifugation (12,000g for 10 min), the supernatants were subjected to total protein assay using the Bradford dye-binding assay (Bio-Rad) within compatible urea concentration (<6 M) by serial dilutions. Contributions of urea to the developed absorbance of the solutions were corrected by adding the same amount of urea to the BSA standard. The total protein solubilized was plotted versus the concentration of urea, and the midpoint of urea solubilization transition region was calculated by complex sigmoid nonlinear analysis (Kim et al. 2001).
Acknowledgments
We thank Prof. B.H. Oh (Pohang University of Science and Technology) and Dr. J.S. Kim (KRIBB) for the gift of plasmids encoding GNBPs and PTPRS and DUSP7, respectively. We also thank Prof. Do-Hyun Jo (Ajou University) for allowing us to use a FPLC system and Yun-Sun Jang for excellent technical assistance in protein purifications. This work was supported by grants from Korea Research Foundation (KRF-2004-003-D00102) and Ajou University to Y.S.K.
Abbreviations
HHP, high hydrostatic pressure
IB, inclusion body
SEC, size-exclusion chromatography
GNBP, Gram-negative binding protein
DTT, dithiothreitol
GSSG, oxidized glutathione
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051813506.
References
- Arakawa, T. and Tsumoto, K. 2003. The effects of arginine on refolding of aggregated proteins: Not facilitate refolding, but suppress aggregation. Biochem. Biophys. Res. Commun. 304: 148–152. [DOI] [PubMed] [Google Scholar]
- Armstrong, N., de Lencastre, A., and Gouaux, E. 1999. A new protein folding screen: Application to the ligand binding domains of a glutamate and kainate receptor and to lysozyme and carbonic anhydrase. Protein Sci. 8: 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, D. and Khanna, N. 1996. Method for increasing the yield of properly folded recombinant human γ interferon from inclusion bodies. J. Biotechnol. 52: 127–133. [DOI] [PubMed] [Google Scholar]
- Baynes, B.M. and Trout, B.L. 2004. Rational design of solution additives for the prevention of protein aggregation. Biophys. J. 87: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynes, B.M., Wang, D.I., and Trout, B.L. 2005. Role of arginine in the stabilization of proteins against aggregation. Biochemistry 44: 4919–4925. [DOI] [PubMed] [Google Scholar]
- Bowden, G.A., Paredes, A.M., and Georgiou, G. 1991. Structure and morphology of protein inclusion bodies in Escherichia coli. Biotechnology 9: 725–730. [DOI] [PubMed] [Google Scholar]
- De Bernardez Clark, E., Hevehan, D., Szela, S., and Maachupalli-Reddy, J. 1998. Oxidative renaturation of hen egg-white lysozyme. Folding vs. aggregation. Biotechnol. Prog. 14: 47–54. [DOI] [PubMed] [Google Scholar]
- De Bernardez Clark, E., Schwarz, E., and Rudolph, R. 1999. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 309: 217–236. [DOI] [PubMed] [Google Scholar]
- Ferrao-Gonzales, A.D., Souto, S.O., Silva, J.L., and Foguel, D. 2000. The preaggregated state of an amyloidogenic protein: Hydrostatic pressure converts native transthyretin into the amyloidogenic state. Proc. Natl. Acad. Sci. 97: 6445–6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foguel, D., Suarez, M.C., Ferrao-Gonzales, A.D., Porto, T.C., Palmieri, L., Einsiedler, C.M., Andrade, L.R., Lashuel, H.A., Lansbury, P.T., Kelly, J.W., et al. 2003. Dissociation of amyloid fibrils of α-synuclein and transthyretin by pressure reveals their reversible nature and the formation of water-excluded cavities. Proc. Natl. Acad. Sci. 100: 9831–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovits, B.M. and Horowitz, P.M. 1998. High hydrostatic pressure can reverse aggregation of protein folding intermediates and facilitate acquisition of native structure. Biochemistry 37: 6132–6135. [DOI] [PubMed] [Google Scholar]
- Gross, M. and Jaenicke, R. 1994. Proteins under pressure. The influence of high hydrostatic pressure on structure, function and assembly of proteins and protein complexes. Eur. J. Biochem. 221: 617–630. [DOI] [PubMed] [Google Scholar]
- Hawley, S.A. 1971. Reversible pressure–temperature denaturation of chymotrypsinogen. Biochemistry 10: 2436–2442. [DOI] [PubMed] [Google Scholar]
- Ho, J.G. and Middelberg, A.P. 2004. Estimating the potential refolding yield of recombinant proteins expressed as inclusion bodies. Biotechnol. Bioeng. 87: 584–592. [DOI] [PubMed] [Google Scholar]
- Ishibashi, M., Tsumoto, K., Tokunaga, M., Ejima, D., Kita, Y., and Arakawa, T. 2005. Is arginine a protein-denaturant? Protein Expr. Purif. 42: 1–6. [DOI] [PubMed] [Google Scholar]
- Jones, L.S., Randolph, T.W., Kohnert, U., Papadimitriou, A., Winter, G., Hagmann, M.L., Manning, M.C., and Carpenter, J.F. 2001. The effects of Tween 20 and sucrose on the stability of anti-L-selectin during lyophilization and reconstitution. J. Pharm. Sci. 90: 1466–1477. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Ryu, J.H., Han, S.J., Choi, K.H., Nam, K.B., Jang, I.H., Lemaitre, B., Brey, P.T., and Lee, W.J. 2000. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and β-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J. Biol. Chem. 275: 32721–32727. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Cape, S.P., Chi, E., Raffen, R., Wilkins-Stevens, P., Stevens, F.J., Manning, M.C., Randolph, T.W., Solomon, A., and Carpenter, J.F. 2001. Counteracting effects of renal solutes on amyloid fibril formation by immunoglobulin light chains. J. Biol. Chem. 276: 1626–1633. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Randolph, T.W., Seefeldt, M.B., and Carpenter, J.F. 2005. High pressure studies on protein aggregates and amyloid fibrils. Methods Enzymol. (in press). [DOI] [PubMed]
- Kreilgaard, L., Jones, L.S., Randolph, T.W., Frokjaer, S., Flink, J.M., Manning, M.C., and Carpenter, J.F. 1998. Effect of Tween 20 on freeze-thawing- and agitation-induced aggregation of recombinant human factor XIII. J. Pharm. Sci. 87: 1597–1603. [DOI] [PubMed] [Google Scholar]
- Kyte, J. and Doolittle, R.F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Lefebvre, B.G. and Robinson, A.S. 2003. Pressure treatment of tailspike aggregates rapidly produces on-pathway folding intermediates. Biotechnol. Bioeng. 82: 595–604. [DOI] [PubMed] [Google Scholar]
- Middelberg, A.P. 2002. Preparative protein refolding. Trends Biotechnol. 20: 437–443. [DOI] [PubMed] [Google Scholar]
- Montalibet, J., Skorey, K.I., and Kennedy, B.P. 2005. Protein tyrosine phosphatase: Enzymatic assays. Methods 35: 2–8. [DOI] [PubMed] [Google Scholar]
- Morais, A.C., Chapeaurouge, A., and Ferreira, S.T. 2005. Acid- and pressure-induced (un)folding of yeast glutathione reductase: Competition between protein oligomerization and aggregation. Int. J. Biochem. Cell. Biol. 37: 1890–1899. [DOI] [PubMed] [Google Scholar]
- Muda, M., Boschert, U., Dickinson, R., Martinou, J.C., Martinou, I., Camps, M., Schlegel, W., and Arkinstall, S. 1996. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J. Biol. Chem. 271: 4319–4326. [DOI] [PubMed] [Google Scholar]
- Randolph, T.W., Seefeldt, M., and Carpenter, J.F. 2002. High hydrostatic pressure as a tool to study protein aggregation and amyloidosis. Biochim. Biophys. Acta 1595: 224–234. [DOI] [PubMed] [Google Scholar]
- Reddy, K.R., Lilie, H., Rudolph, R., and Lange, C. 2005. L-Arginine increases the solubility of unfolded species of hen egg white lysozyme. Protein Sci. 14: 929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royet, J., Reichhart, J.M., and Hoffmann, J.A. 2005. Sensing and signaling during infection in Drosophila. Curr. Opin. Immunol. 17: 11–17. [DOI] [PubMed] [Google Scholar]
- Sackett, D.L. 1997. Natural osmolyte trimethylamine N-oxide stimulates tubulin polymerization and reverses urea inhibition. Am. J. Physiol. 273: R669–R676. [DOI] [PubMed] [Google Scholar]
- Seefeldt, M.B., Ouyang, J., Froland, W.A., Carpenter, J.F., and Randolph, T.W. 2004. High-pressure refolding of bikunin: Efficacy and thermodynamics. Protein Sci. 13: 2639–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki, K., Kudou, M., Fujiwara, S., Imanaka, T., and Takagi, M. 2002. Biophysical effect of amino acids on the prevention of protein aggregation. J. Biochem. 132: 591–595. [DOI] [PubMed] [Google Scholar]
- Silva, J.L. and Weber, G. 1993. Pressure stability of proteins. Annu. Rev. Phys. Chem. 44: 89–113. [DOI] [PubMed] [Google Scholar]
- St. John, R.J., Carpenter, J.F., and Randolph, T.W. 1999. High pressure fosters protein refolding from aggregates at high concentrations. Proc. Natl. Acad. Sci. 96: 13029–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. John, R.J., Carpenter, J.F., Balny, C., and Randolph, T.W. 2001. High pressure refolding of recombinant human growth hormone from insoluble aggregates. Structural transformations, kinetic barriers, and energetics. J. Biol. Chem. 276: 46856–46863. [DOI] [PubMed] [Google Scholar]
- St. John, R.J., Carpenter, J.F., and Randolph, T.W. 2002. High-pressure refolding of disulfide-cross-linked lysozyme aggregates: Thermodynamics and optimization. Biotechnol. Prog. 18: 565–571. [DOI] [PubMed] [Google Scholar]
- Timasheff, S.N. 1998. Control of protein stability and reactions by weakly interacting cosolvents: The simplicity of the complicated. Adv. Protein Chem. 51: 355–432. [DOI] [PubMed] [Google Scholar]
- Van Eldik, R., Asano, T., and Le Noble, W. 1989. Activation and reaction volumes in solution. 2. Chem. Rev. 89: 549–688. [DOI] [PubMed] [Google Scholar]
- Wagner, J., Gordon, L.A., Heng, H.H., Tremblay, M.L., and Olsen, A.S. 1996. Physical mapping of receptor type protein tyrosine phosphatase sigma (PTPRS) to human chromosome 19p13.3. Genomics 38: 76–78. [DOI] [PubMed] [Google Scholar]
- Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H., and Hultmark, D. 2000. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. 97: 13772–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetlaufer, D.B. and Xie, Y. 1995. Control of aggregation in protein refolding: A variety of surfactants promote renaturation of carbonic anhydrase II. Protein Sci. 4: 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, R. and Dzwolak, W. 2005. Exploring the temperature-pressure configurational landscape of biomolecules: From lipid membranes to proteins. Philos. Transact. A Math. Phys. Eng. Sci. 363: 537–562; discussion 562–563. [DOI] [PubMed] [Google Scholar]
- Yasuda, M., Murakami, Y., Sowa, A., Ogino, H., and Ishikawa, H. 1998. Effect of additives on refolding of a denatured protein. Biotechnol. Prog. 14: 601–606. [DOI] [PubMed] [Google Scholar]