Figure 1.
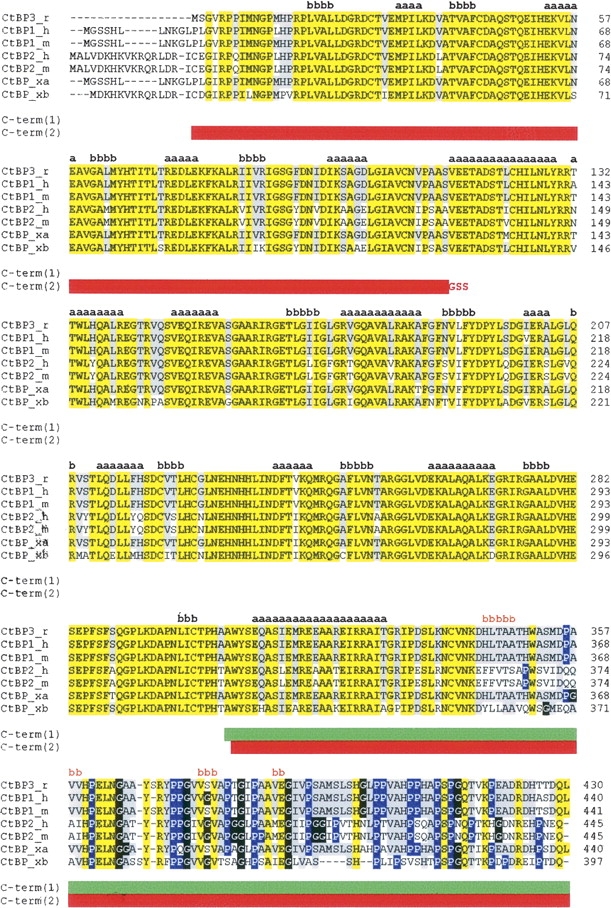
CtBP sequence alignments. Rat CtBP3 (CtBP3_r, SWISS-PROT:09Z2F5) is aligned with human CtBP1 (CtBP1_h, SWISS-PROT:Q13363), mouse CtBP1 (CtBP1_m, SWISS-PROT:088712), human CtBP2 (CtBP2_h, SWISS-PROT:P56545), mouse CtBP2 (CtBP2_m, SWISS-PROT:P56546), and Xenopus laevis CtBP (isoforms CtBP_xa, SWISS-PROT:Q9YHU; CtBP_xb, SWISS-PROT:Q9W758). The sequence alignment was performed using the CLUSTALW program (http://www.ebi.ac.uk/clustalw). Residues identical in all sequences are highlighted in yellow. Residues identical to CtBP3 in >50% of the aligned sequences are highlighted in gray. Pro and Gly residues in the C-terminal region are shown in blue and in black, respectively. Secondary structure of t-CtBP3 (residues 15–345) and secondary structure prediction for the C-terminal region are shown in black and in orange, respectively, with a = helix and b = strand. The C-term(1) and C-term(2) constructs are indicated by green and by red horizontal bars, respectively.
