Figure 8.
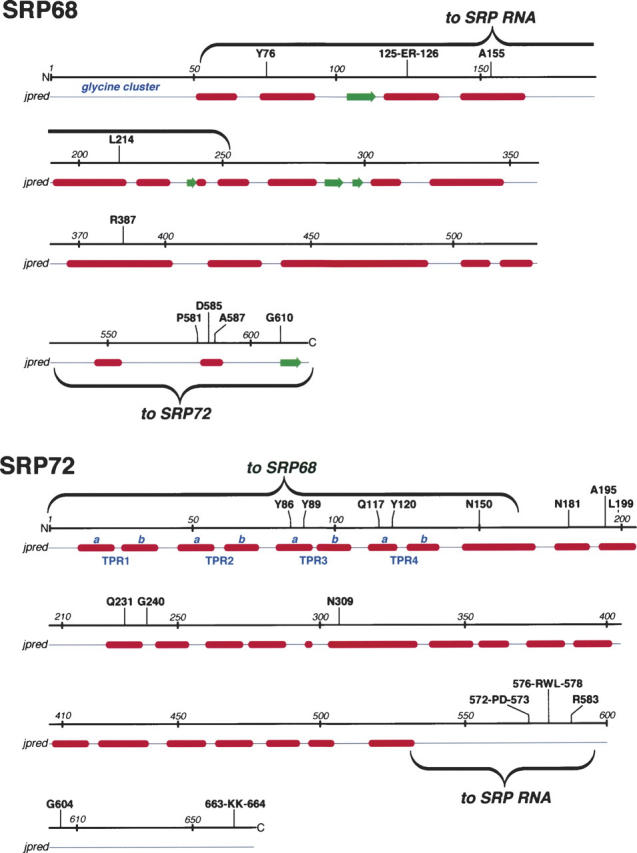
Features of SRP68/72. For both proteins, amino acid positions are given in increments of ∼50 residues. Amino acid residues that are invariant or highly conserved in the multiple sequence alignments (provided as Supplemental Material Sup1-SRP68.pdf and Sup2-SRP72.pdf) are shown above the lines. Helices (red) and β-structures (green) as predicted by jpred (Cuff and Barton 2000) are shown below the lines. Annotated in blue are the glycine cluster at the N terminus of SRP68 and the four predicted TPR-like motifs (Blatch and Lassle 1999) with their divisions into helices a and b. The wavy brackets designate the regions responsible for binding to SRP RNA and for the binding of the two proteins to each other.
