Figure 1.
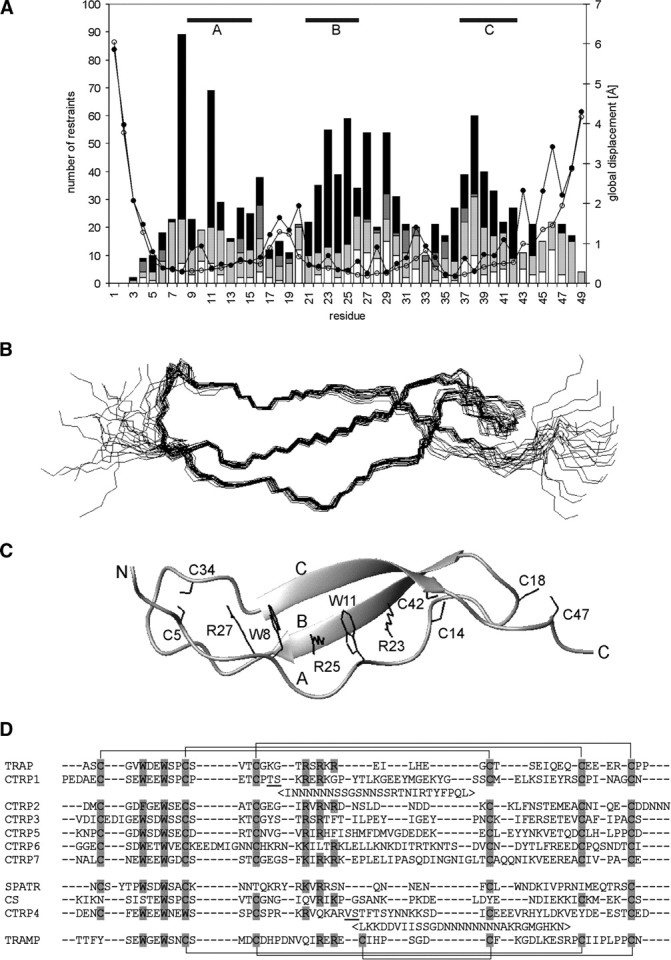
Structure of TRAP-TSR. (A) Number of NOE restraints [(black) long range, i − j ≥ 5; (dark gray) medium range, i − j < 5; (light gray) sequential, i − j = 1; (white) intraresidual, i − j = 0] and RMSD [(open circles) backbone atoms; (filled circles) all heavy atoms] per residue. The areas of regular structure are shown above. (B) Ensemble of 20 energy minimized structures of TRAP-TSR domain; (C) secondary structure together with the stabilizing stack of tryptophan, arginine, and cysteine side chains shown specifically. Protons are omitted from the figure for reasons of clarity. The three strands and the two termini are marked with capital letters. (D) Sequence alignment of P. falciparum protein domains reported to have sequence similarity to TSRs. The proteins are TRAP, CTRP (CS-protein-TRAP-related protein; Trottein et al. 1995) with seven TSR domains, SPATR (Chattopadhyay et al. 2003), CS (Dame et al. 1984), and TRAMP (thrombospondin-related apical merozoite protein; Thompson et al. 2004). TRAP and six of the CTRP TSRs have their six cysteines organized in the manner typical for Group 2 TSRs. TRAMP TSR belongs to Group 1 TSRs. The potential locations of disulfide bonds are shown above the alignment (Group 2) and below it (Group 1). SPATR, CS, and CTRP4 have a distinct pattern of conserved residues and possibly divergent structures. Potential disulfide and stack forming residues are highlighted. CTRP1 and CTRP4 have long insertions between the underlined residues. The location of these insertions is as in Tan et al. (2002). The alignment was made with ClustalW (1.83) by excluding the insertions.
