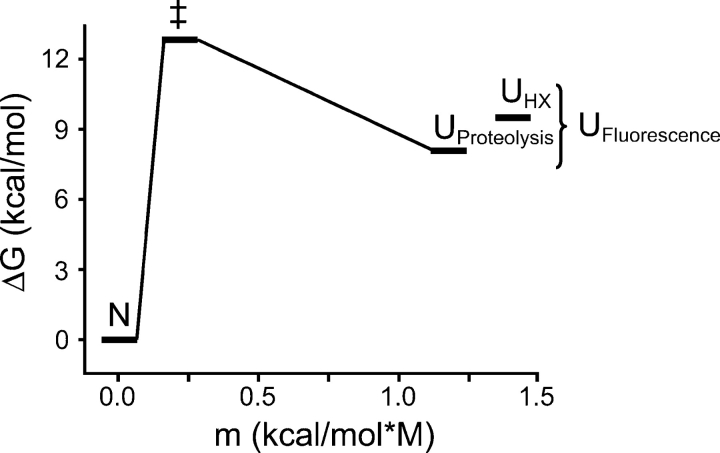
Figure 6.
Experimentally determined energy landscape of Src SH2. The energy of each state is given relative to the native state N. The reaction coordinate is defined by the m-value of each transition, relative to N, a measure of the solvent-accessible surface area exposed in the transition. ΔG‡ is calculated using a preexponential factor of 108 sec−1, estimated from intrachain triplet energy transfer rates in unfolded peptides (Krieger et al. 2003). N, native state; ‡, transition state for (un)folding; Uproteolysis, unfolded state detected by proteolysis; and UHX, unfolded state detected by hydrogen exchange. The fluorescence signal does not distinguish between the two unfolded states. The height of the barrier, if any, connecting the two unfolded states has not been determined.