Figure 1.
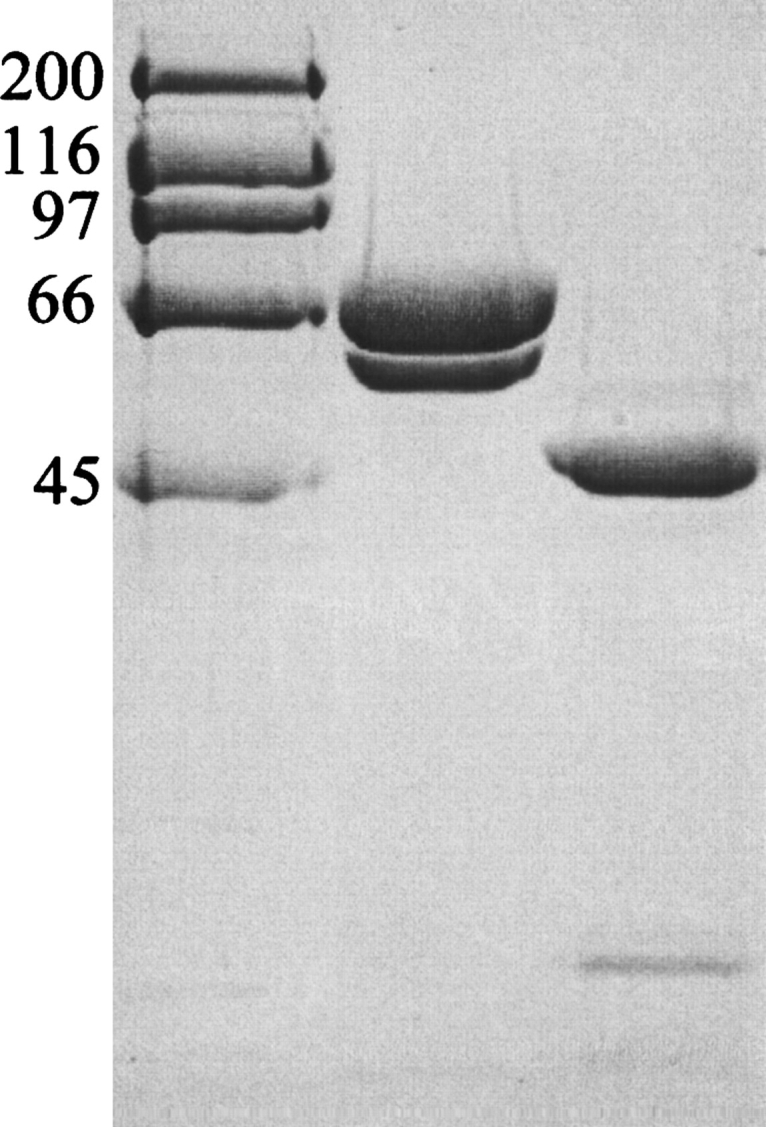
SDS-PAGE of truncated PBP1b protein samples. A 10% (w/v) acrylamide separating gel was used, with subsequent steps following standard procedure. (Lane 1) High-range molecular markers (Bio-Rad; molecular weight given in kilodaltons). (Lane 2) “Freshly prepared” truncated PBP1b protein stock. (Lane 3) Moenomycin-aged truncated PBP1b protein stocks. The aging of protein samples was carried out in an attempt to mimic any proteolysis occurring in the crystallization drop. N-terminal protein sequencing results were used to characterize the protein bands: Lane 2, upper band MD85KVRV, lower band L184IKQQV; lane 3, upper band Q323DFLPS, lower band K139AIIAT. The desired soluble truncation product of our PBP1b construct was present in freshly prepared protein samples, starting at the native sequence residue D85 (post-transmembrane helix). This sample also contains a contaminating band starting at L184, resulting from proteolysis of the GT domain. After protein aging, the larger, two-domain constructs are pared down to the linker region and TP domain (Q323 onward). The smaller, TP-associated hairpin from the GT domain (to the smallest possible extent, residues 105–119) is not visible on this gel. If the degradation observed in aged samples is comparable to that in the crystallization drop, residues 323–336 must be disordered in the electron density map. It is also possible that further proteolysis has occurred precrystallization at the protease-sensitive R336 position (Macheboeuf et al. 2005).
