Abstract
l-Colitose is a 3,6-dideoxysugar found in the O-antigens of some Gram-negative bacteria such as Escherichia coli and in marine bacteria such as Pseudoalteromonas tetraodonis. The focus of this investigation, GDP-4-keto-6-deoxy-d-mannose-3-dehydratase, catalyzes the third step in colitose production, which is the removal of the hydroxyl group at C3′ of GDP-4-keto-6-deoxymannose. It is an especially intriguing PLP-dependent enzyme in that it acts as both a transaminase and a dehydratase. Here we present the first X-ray structure of this enzyme isolated from E. coli Strain 5a, type O55:H7. The two subunits of the protein form a tight dimer with a buried surface area of ∼5000 Å2. This is a characteristic feature of the aspartate aminotransferase superfamily. Although the PLP-binding pocket is formed primarily by one subunit, there is a loop, delineated by Phe 240 to Glu 253 in the second subunit, that completes the active site architecture. The hydrated form of PLP was observed in one of the enzyme/cofactor complexes described here. Amino acid residues involved in anchoring the cofactor to the protein include Gly 56, Ser 57, Asp 159, Glu 162, and Ser 183 from one subunit and Asn 248 from the second monomer. In the second enzyme/cofactor complex reported, a glutamate ketimine intermediate was found trapped in the active site. Taken together, these two structures, along with previously reported biochemical data, support the role of His 188 as the active site base required for catalysis.
Keywords: PLP-dependent enzyme; colitose; molecular structure; 3,6-dideoxysugars; O-antigens
The biochemical importance of carbohydrates cannot be overstated, for they are essential elements in nearly every physiological process and represent the most abundant biomolecules in living systems. Di- and trideoxysugars are an especially important and intriguing class of carbohydrates that are synthesized by plants, fungi, and bacteria (Kennedy and White 1983; Rupprath et al. 2005). Many are formed from simple monosaccharides such as glucose 6-phosphate or fructose 6-phosphate via a myriad of enzymatic reactions including dehydrations, reductions, and methylations. Most of the deoxysugars of biological relevance are constructed around the 3,6-dideoxyhexoses or the 2,3(4),6-trideoxyhexoses, which are further modified by epimerization, amination, and C-, N-, and O-methylation reactions. These modifications yield an astonishing array of biological molecules with differing three-dimensional shapes and hydrophilicities.
Several of these unusual sugars have been isolated from the O-antigens of various Gram-negative bacteria, where they contribute to the enormous structural variations observed in bacterial cell walls (Lerouge and Vanderleyden 2002). Others have been found in polyketide antibiotics such as erythromycin, where they have been shown to provide or enhance the pharmacological activities of the respective drugs (Weymouth-Wilson 1997).
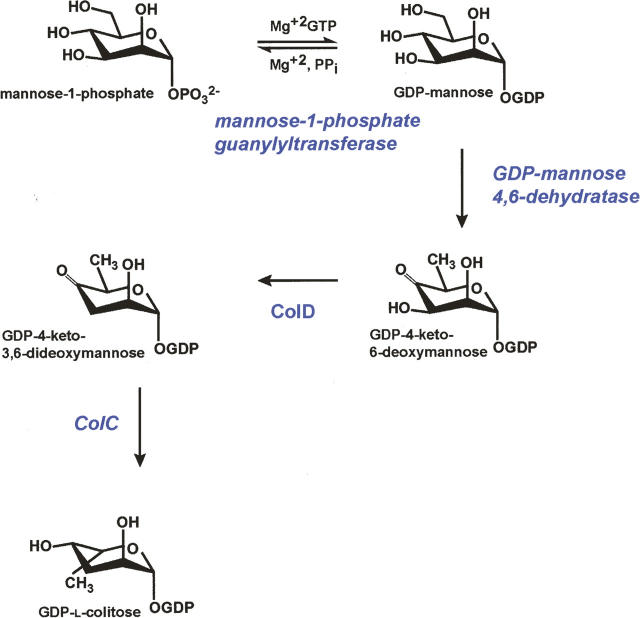
One of these unusual sugars is l-colitose (Scheme 1), a 3,6-dideoxysugar isolated from the O-antigens of Gram-negative bacteria such as Escherichia coli (Edstrom and Heath 1965), Yersinia pseudotuberculosis (Kotandrova et al. 1989), Salmonella enterica (Xiang et al. 1993), Vibrio cholerae (Hisatsune et al. 1993), and in marine bacteria such as Pseudoalteromonas tetraodonis (Muldoon et al. 2001) and Pseudoalteromonas carrageenovora (Silipo et al. 2005). Reports describing the isolation of colitose date as far back as 1958 (Luderitz et al. 1958).
Scheme 1.
The biosynthesis of the O-antigen in bacteria requires nucleotide-linked sugars, which are transferred to the growing oligosaccharide chain. Four enzymes are involved in the production of GDP-l-colitose, as outlined in Scheme 1 (Bastin and Reeves 1995; Beyer et al. 2003). The focus of this investigation is GDP-4-keto-6-deoxy-d-mannose-3-dehydratase, which catalyzes the third step in the pathway, namely the removal of the hydroxyl group at C3′ of the hexose. The enzyme is encoded by the colD gene, and hereafter is referred to as ColD. It is an especially intriguing PLP-dependent enzyme in that it lacks the conserved lysine that typically anchors the cofactor to the protein. Recent biochemical studies have demonstrated that the enzyme from Y. pseudotuberculosis is bifunctional, with both deoxygenation and transamination activities (Beyer et al. 2003; Alam et al. 2004), and as such, it represents a new paradigm for unusual deoxysugar formation.
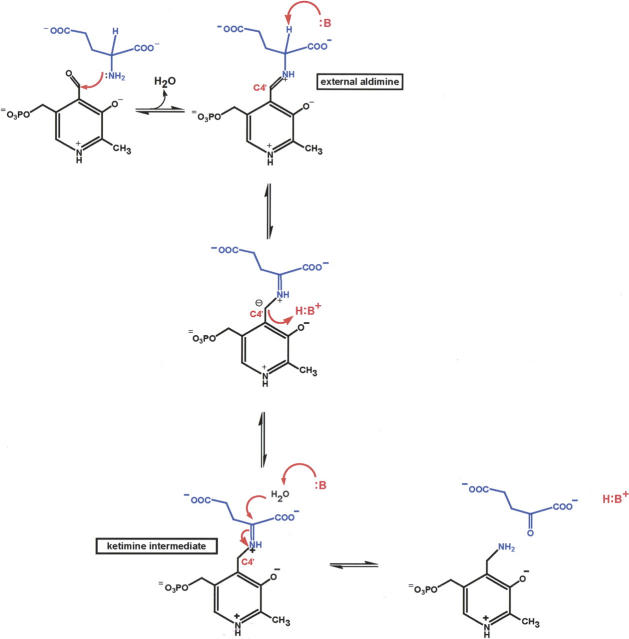
PLP (pyridoxal 5′-phosphate) is a remarkable and versatile coenzyme that is ubiquitous in nature. Enzymes dependent upon PLP catalyze a wide range of chemical reactions including decarboxylations, racemizations, transaminations, and β-eliminations, among others (Jansonius 1998; Toney 2005). Most PLP-dependent enzymes contain a conserved lysine residue, which forms a Schiff base with the coenzyme in the resting state. This species is referred to as the internal aldimine. In ColD, an internal aldimine is not possible due to the replacement of a histidine for the conserved lysine. Rather, in ColD, PLP is first converted to pyridoxamine 5′-phosphate (PMP) by reacting with glutamate as indicated in Scheme 2. In the first step, glutamate forms a Schiff base with the noncovalently bound PLP, a species referred to as the external aldimine. Subsequently, an enzymatic base deprotonates the aldimine intermediate, leading to the formation of a carbanion that is stabilized by various resonance forms. Protonation of the carbanion at C4′ of the PLP by an active site acid leads to a ketimine intermediate which, upon hydrolysis, results in PMP and α-ketoglutarate.
Scheme 2.
Here we describe the three-dimensional structures of ColD from Escherichia coli, Strain 5a, type O55:H7, with a bound hydrated form of PLP and with a trapped glutamate ketimine intermediate. The employment of PMP in a dehydration reaction is quite rare, and thus these structures provide a wealth of new molecular insight into this novel coenzyme B6-dependent enzyme.
Results
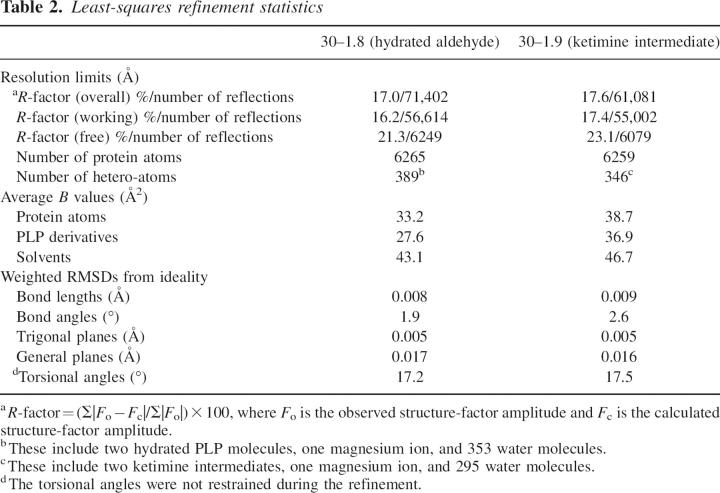
The first structure of ColD was determined to 1.8 Å resolution. For this structural analysis, crystals were grown in the presence of PLP and 2 mM α-ketoglutarate. As can be seen in Figure 1A, the two subunits of ColD form a tight dimer with an extensive total buried surface area of ∼5000 Å2, based on a probe radius of 1.4 Å (Zhang and Matthews 1995). This type of quaternary structure is characteristic of the aspartate aminotransferase superfamily (Jansonius 1998). The dimer has overall dimensions of ∼80 Å × 70 Å × 70 Å, with the active sites separated by ∼20 Å. Electron density corresponding to the entire polypeptide chain backbone is continuous from the N to C termini for both monomers in the asymmetric unit. The conformations of the two subunits forming the dimer are virtually identical, and correspond with a root-mean-square deviation (RMSD) of 0.14 Å for 367 structurally equivalent α-carbons.
Figure 1.
The structure of ColD. The crystals of ColD employed in this investigation contained a dimer in the asymmetric unit as shown in A. The subunits are related by a twofold rotational axis, which lies perpendicular to the plane of the page. The PLP cofactors, separated by ∼21 Å, are displayed in space-filling representations. For all figures, the coordinates corresponding to subunits 1 and 2 of the dimer are color coded in blue and green, respectively. A separate ribbon representation of subunit 2 is given in B. The 10 β-strands characterizing the subunit are labeled A–J. Part of the active site region for subunit 2 is formed by a loop contributed by subunit 1 (Phe 240–Glu 253) as highlighted in C.
A ribbon representation of an individual subunit (a total of 388 amino acids) is displayed in Figure 1B. It has a somewhat concave-shaped appearance with molecular dimensions of ∼60 Å × 65 Å × 70 Å. The polypeptide chain initiates with two α-helices defined by Asp 13–Ser 25 and Glu 32–Phe 45. These helices lead up to the first strand of β-sheet labeled “A” in Figure 1B. Overall, the fold of the subunit is dominated by a mixed eight-stranded β-sheet flanked by eight α-helices. The β-strands are labeled “A,” “G,” “F,” “E,” “D,” “B,” “C,” and “I” in Figure 1B, and correspond to Tyr 49–Val 53, Gly 196–Thr 200, Met 179–Ser 184, Ile 155–Asp 159, Lys 128–Asn 134, Glu 80–Pro 84, Arg 101–Val 105, and Thr 351–His 353, respectively. Note that β-strands A, G, and F run antiparallel. In addition to this large β-sheet, there are two strands of antiparallel sheet labeled “H” and “J” that correspond to residues Gly 303–Ile 308, and Leu 367–Val 369, respectively. A rather large helix, formed by Glu 253–Lys 283, packs against both the first helix at the N terminus and the last helix at the C terminus. The third helix of the subunit, which connects β-strands A and B, is positioned such that the positive end of its helix dipole moment projects toward the phosphate of the PLP cofactor. Those residues involved in anchoring the PLP ligand to the protein are provided primarily by one subunit. However, as shown in Figure 1C, there is a loop, delineated by Phe 240–Glu 253, which forms one wall of the active site and is provided by the second subunit of the dimer.
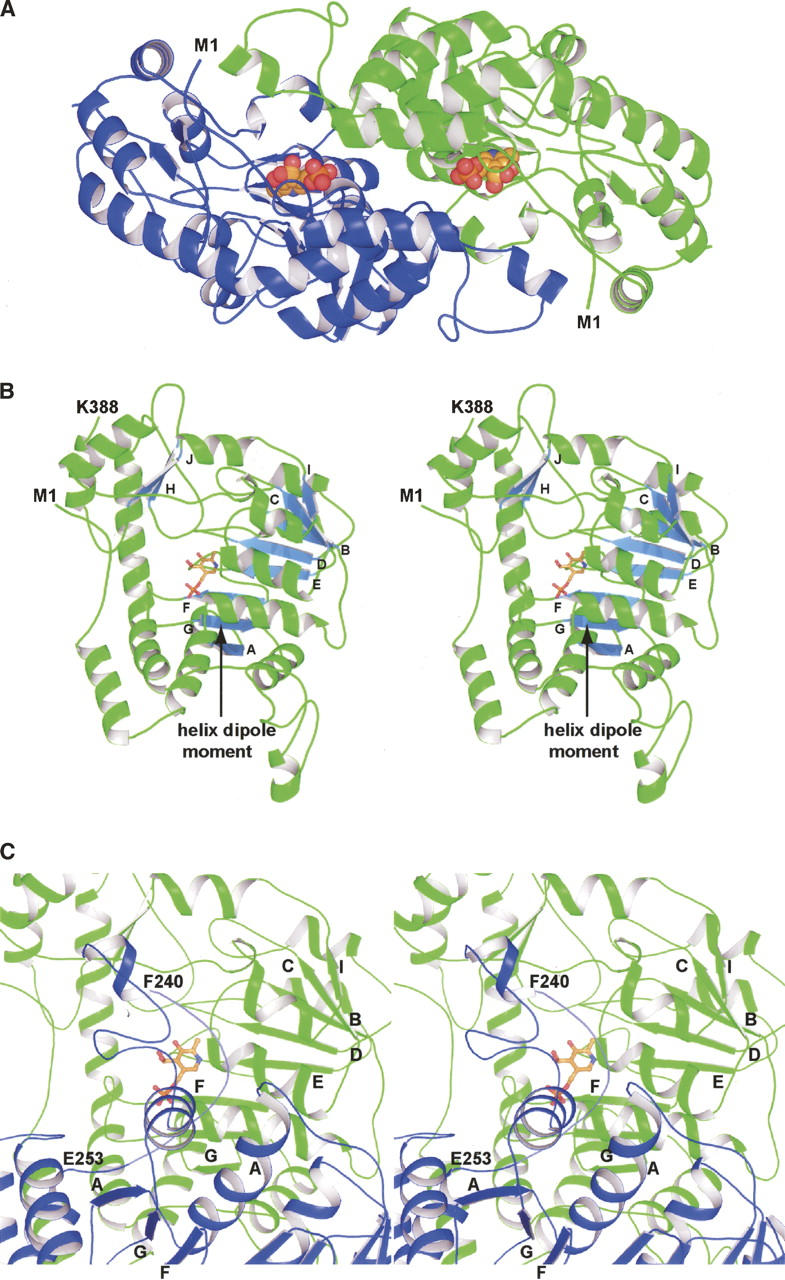
Unlike the typical PLP-dependent enzymes, ColD does not contain the lysine residue necessary to form a Schiff base between its side chain Nζ and C4′ of the cofactor (the internal aldimine). Aldehydes, like PLP, react with water to form hydrates, as indicated in Figure 2A. Whether the carbonyl or hydrate moiety is favored at equilibrium depends upon the chemical nature of the aldehyde (or ketone). In the case of free PLP in solution, the aldehyde form is favored. There is spectral evidence, however, that when PLP is bound to a protein, the hydrate may be favored (Harris et al. 1976; Nishino et al. 1997; Tramonti et al. 2002). Shown in Figure 2B is the electron density corresponding to the PLP ligand in subunit 2. Originally the ligand was modeled into the electron density as an aldehyde with C4′ having sp2 hybridization. As the model refinement continued, however, it became clear from electron density maps calculated with (Fo − Fc) coefficients that, in fact, the hydrated form of PLP, with C4′ having sp3 hybridization, was trapped in the active site.
Figure 2.
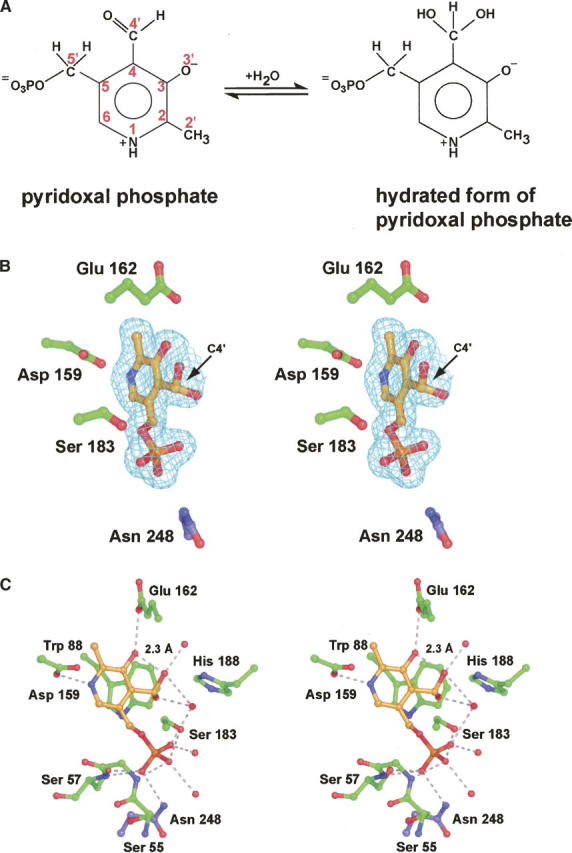
The complex of ColD with the hydrated form of PLP. Aldehydes react with water to form hydrates. This type of reaction for PLP is shown in A, along with the numbering used to define the various atoms of the cofactor. Electron density corresponding to the hydrated form of PLP is displayed in B. The map, contoured at 2σ, was calculated with coefficients of the form (Fo − Fc), where Fo was the native structure factor amplitude and Fc was the calculated structure factor amplitude from the model lacking coordinates for the PLP. Those residues situated within ∼3.5 Å of the hydrated PLP in subunit 2 are shown in C. The dashed lines indicate possible hydrogen bonds between atoms positioned within 3.2 Å of each other. Asn 248 is contributed by subunit 1.
Those residues located within ∼3.5 Å of the hydrated PLP in subunit 2 are shown in Figure 2C. Most of the protein:ligand interactions occur near the phosphoryl group. Indeed, the phosphoryl moiety is anchored firmly in place by eight hydrogen bonding interactions. Specifically, the backbone amide groups of Gly 56 and Ser 57 participate in hydrogen bonding interactions with two of the phosphoryl oxygens of PLP. In addition, the side chain atoms, Oγ of Ser 57 (subunit 2) and Nδ2 of Asn 248 (subunit 1), lie within 2.4 Å and 2.9 Å, respectively, of one of the phosphoryl oxygens. Ser 183 provides another hydrogen bonding interaction between its side chain Oγ and a PLP phosphoryl oxygen. Three ordered water molecules complete the hydrogen bonding pattern observed around the phosphoryl group. Only two side chains, Asp 159 and Glu 162, form hydrogen bonds with the pyridoxal ring of PLP. Asp 159 is positioned at the end of β-strand E, whereas Glu 162 resides in a Type I turn. For the side chain oxygen of Glu 162 to lie within ∼2.8 Å of O3′ of the pyridoxal ring, one of these oxygens must be protonated. The indole ring of Trp 88 provides an off-centered parallel stacking interaction with the cofactor. Note that His 188, which is a lysine residue in most PLP-dependent enzymes, is located at ∼4 Å from the cofactor C4′.
For the second structure reported here, ColD was crystallized in the presence of 2 mM glutamate and the structure solved to 1.9 Å resolution. Again, the two subunits in the asymmetric unit are virtually identical, and correspond with an RMSD of 0.18 Å for 372 structurally equivalent α-carbons.
A close-up view of the electron density observed in the active site of subunit 2 is displayed in Figure 3A. Initially the external aldimine form of PLP was modeled into the electron density whereby the α-amino nitrogen of glutamate and C4′ of the pyridoxal ring form a Schiff base. For this intermediate, the hybridization about the α-carbon of glutamate is sp3 while the hybridization about C4′ of the cofactor is sp2 (Scheme 2). Accordingly, the bond angles about the α-carbon and C4′ should be ∼109° and ∼120°, respectively. During the course of the structure refinement, however, the external aldimine model never matched the electron density well. If, however, the hybridization about the α-carbon and C4′ were restrained to sp2 and sp3, respectively, the model fitted strikingly well into the electron density (Fig. 3A). This type of geometry at the α-carbon and C4′ would be expected for the glutamate ketimine intermediate (Scheme 2). While caution must be applied when interpreting electron density maps calculated at 1.9 Å resolution, the observation of such an intermediate is not unprecedented. Indeed, it has been previously reported in the elegant structural analyses of aspartate aminotransferase, which were conducted at 2.3–2.4 Å resolution (Malashkevich et al. 1993). Aspartate aminotransferase contains the typical lysine residue observed in most PLP-dependent enzymes.
Figure 3.
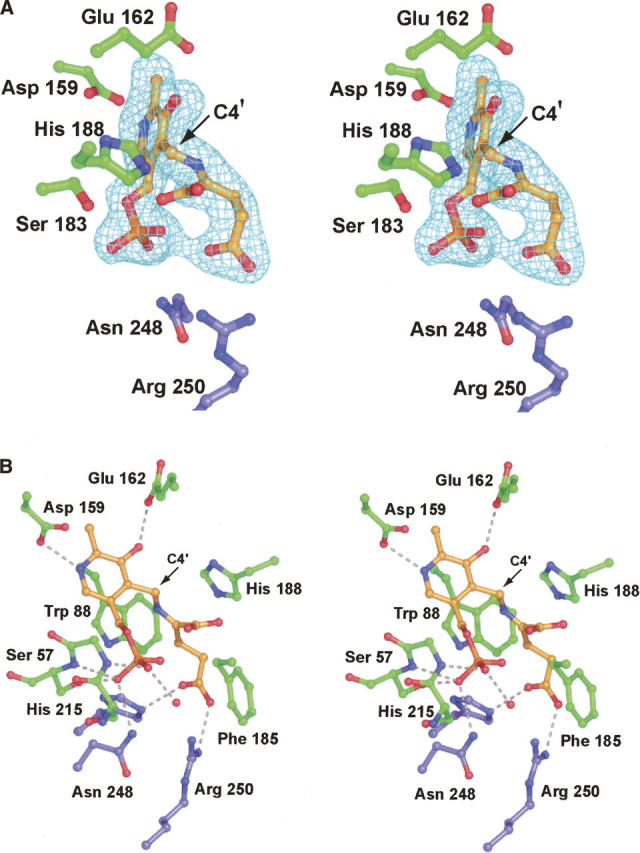
The structure of ColD with a glutamate ketimine intermediate trapped in the active site. Electron density corresponding to the ketimine intermediate is presented in A. The map was calculated in the same manner as described for Figure 2B. Note that the α-carboxylate group of the intermediate is not well ordered, suggesting free rotation. A close-up view of the active site is displayed in B. Possible hydrogen bonding interactions are indicated by the dashed lines. For the sake of clarity, the hydrogen bonding interaction between Ser 183 and the phosphoryl group of the ketimine intermediate was omitted. Ser 183 forms the same interaction with the ketimine phosphoryl group as it does with hydrated PLP (Fig. 2C).
Those residues lying within 3.5 Å of the glutamate ketimine intermediate in subunits 1 and 2 are displayed in Figure 3B. Note that the α-carbon chains for the ColD dimer, whether complexed with either the hydrated or ketimine form of PLP, are virtually identical (RMSD of 0.09 Å). The protein accommodates the more bulky ketimine intermediate by the exclusion of water molecules from the active site rather than by major side chain adjustments. Both His 215 and Arg 250 from the first subunit form electrostatic interactions with the side chain carboxylate group of the ketimine intermediate. As observed for the protein complexed with the PLP hydrate, one of the carboxylate oxygens of Glu 162 lies quite closely to O3′ of the pyridoxal ring (∼2.6 Å), which is indicative of a proton being shared between them.
Discussion
The structure of ColD was solved via molecular replacement with the X-ray coordinates for ArnB from Salmonella typhimurium (Noland et al. 2002). These two enzymes demonstrate a 23% amino acid sequence identity. ArnB plays a key role in lipid A modification by catalyzing the amination at C-4″ of UDP-l-threo-pentapyranosyl-4″-ulose. Both ColD and ArnB are dimeric. Their structurally equivalent α-carbons correspond with an RMSD of 2.5 Å. A superposition of one subunit of ColD onto that of ArnB is displayed in Figure 4A. As can be seen, the overall backbone traces for the two enzymes are very similar. They begin to align at Leu 6 in ColD and Phe 13 in ArnB. The two proteins differ primarily in three areas labeled R1, R2, and R3 in Figure 4A. The first region of difference occurs in the loop defined by Phe 68 to Arg 74, which serves to connect the third α-helix of the ColD subunit and β-strand B. The second area occurs near Lys 224 to Val 230 where, in ColD, these residues adopt a β-hairpin motif. This region is apparently more flexible in ArnB where there is a break in the model from Asp 220 to Gln 232. Finally, the loops leading out of β-strands H in ColD (Lys 309 to Ile 315) and ArnB adopt significantly different orientations (Fig. 4A). In ArnB, Phe 330 is in the cis peptide conformation. The structurally equivalent residue in ColD is Thr 334, which adopts strained dihedral angles (ϕ = ∼58°, ψ = ∼−116°).
Figure 4.
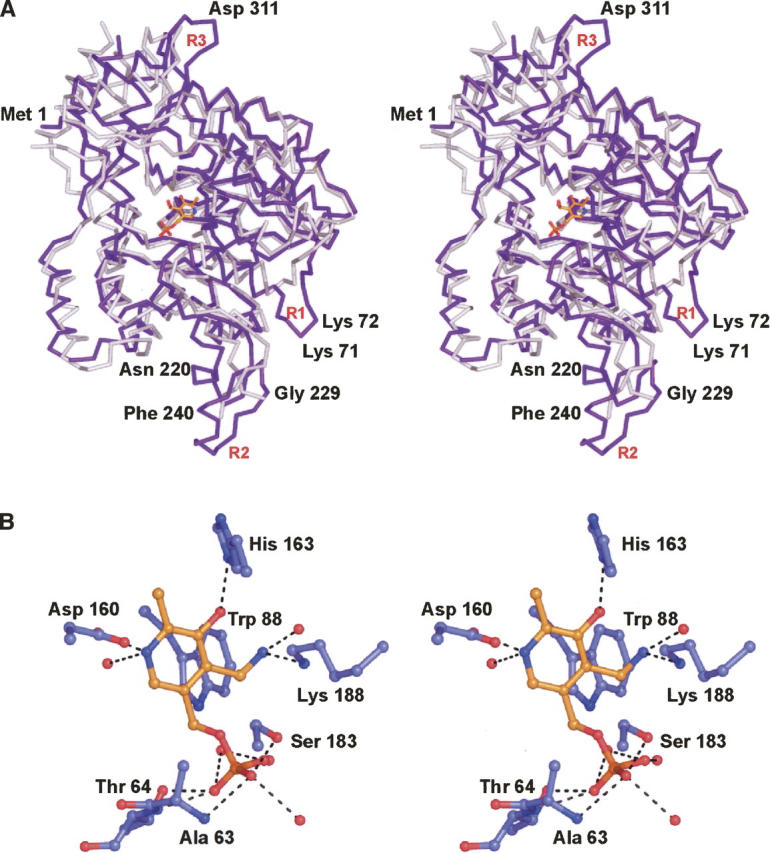
Comparison of ColD with ArnB. A superposition of the α-carbon traces for ColD (purple) and ArnB (white) is presented in A. Labeled residues are for ColD. The observed hydrogen-bonding pattern around PMP in ArnB is shown in B. X-ray coordinates for this figure were obtained from the Protein Data Bank (accession no. 1MDO).
The hydrogen-bonding pattern surrounding the PMP cofactor in ArnB is displayed in Figure 4B. Like that observed in ColD, the phosphate group is anchored to the protein through side chain hydroxyl groups and amide nitrogens and, in addition, is surrounded by several ordered water molecules. The main difference in the binding of PMP to ArnB occurs at His 163, which, in ColD is a glutamate. As observed in ColD, the indole side chain of Trp 88 in ArnB forms a parallel stacking interaction with the pyridoxamine ring of the cofactor.
Once the model for ColD was built and refined, a search with DALI (Holm and Sander 1996) revealed the closest structural relative of ColD to be 3-amino-5-hydroxybenzoic acid synthase (ABHA synthase) rather than ArnB (Eads et al. 1999). ABHA synthase and ColD superimpose with an RMSD of 1.8 Å for 677 structurally equivalent α-carbons. These two proteins display an even lower amino acid sequence identity of 18% amino acid, however. ABHA synthase catalyzes the α,β-dehydration and 1,4-enolization of 5-deoxy-5-amino-3-dehydroshikimic acid to produce 3-amino-5-hydroxybenzoic acid, which serves as the starter unit for the production of ansamycin antibiotics (Ghisalba and Nuesch 1981). A superposition of the polypeptide chains for ColD and ABHA synthase is presented in Figure 5A. As noted for the comparison of the ColD and ArnB models, there are three loops (Phe 68–Arg 74, Lys 224–Val 230, and Lys 309–Ile 315) where ColD and ABHA synthase differ to any significant extent. The cis peptide in ArnB is not conserved in ABHA synthase. A close-up view of the binding region for the internal aldimine in ABHA synthase is depicted in Figure 5B. The hydrogen bonding pattern around the phosphate is a similar to that observed in both ColD and ArnB. As seen in ArnB, a histidine side chain lies within 2.8 Å of O3′ of the pyridoxamine ring. This residue varies among PLP-dependent enzymes, and has also been observed as an asparagine or aspartate. Trp 88 in ColD, which stacks with the pyridoxal ring of the hydrated PLP cofactor, is replaced with a phenylalanine in ABHA synthase.
Figure 5.
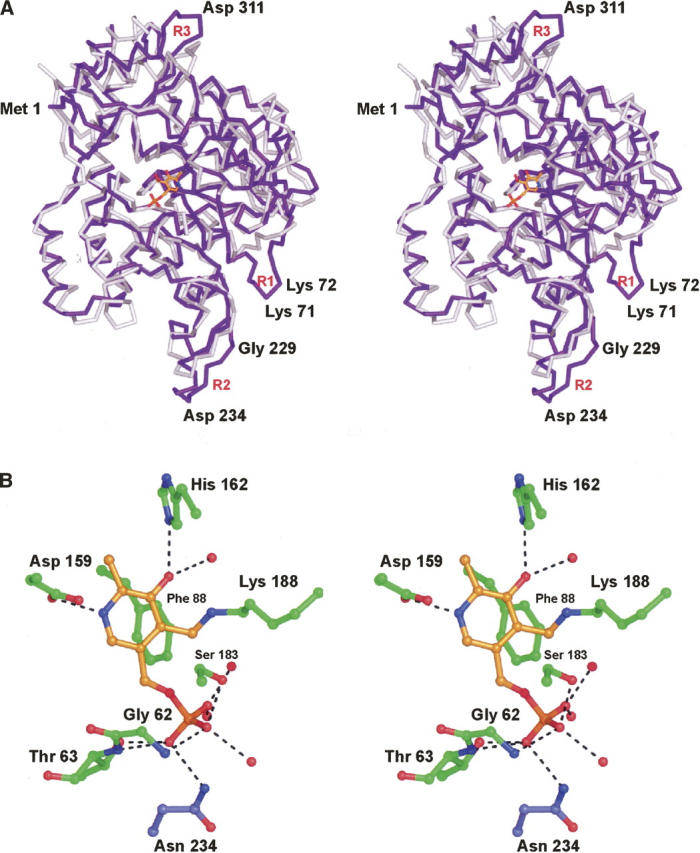
Comparison of ColD with ABHA synthase. Shown in A is a superposition of the α-carbon traces for ColD (purple) and ArnB (white). Labeled residues are for ColD. The observed hydrogen-bonding pattern around the internal aldimine in ABHA synthase is shown in B. X-ray coordinates for this figure were obtained from the Protein Data Bank (accession no. 1B9H). Those residues highlighted in green belong to one subunit, while Asn 234 is contributed by the second subunit in the dimer of ABHA synthase.
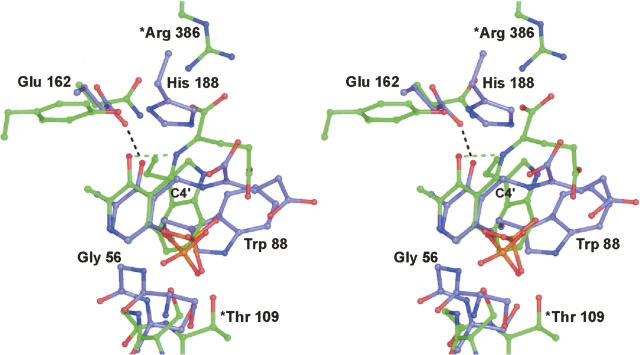
A glutamate ketimine intermediate was first observed structurally in chicken mitochondrial aspartate aminotransferase (AATase) (Malashkevich et al. 1993). The polypeptide chains for the dimers of ColD and AATase superimpose less well with an RMSD of 3.5 Å for 531 structurally equivalent α-carbons. Not surprisingly, there are significant differences in the active sites of these two enzymes. As can be seen in Figure 6, the conformations of the trapped ketimine intermediates in ColD and AATase are quite different, with variations initiating at C4′ of the cofactor. In AATase, the pyridoxamine ring of the cofactor and N, Cα, Cβ, and the α-carboxylate group of glutamate all lie nearly in a plane. Furthermore, the glutamate (or Schiff base) nitrogen sits within 2.5 Å from the pyridoxamine O3′ in the AATase ketimine. This is indicative of a proton being shared between these two atoms. Strikingly, the distance between O3′ and the Schiff base nitrogen in the ColD model is 3.5 Å. Moreover, in ColD, O3′ lies at 2.7 Å from the carboxylate of Glu 162, which means that there is a proton located somewhere between them. In light of these differences, it can be speculated that in the ColD ketimine intermediate a proton is not shared between O3′ and the Schiff base nitrogen as is so often drawn in textbooks, but rather the proton is formally bonded to the nitrogen. This is referred to as the ketoenamine tautomer, and it has been postulated that enzymes can enhance their catalytic activity by promoting this tautomeric form (Toney 2001). The two ketimine intermediates observed in ColD and AATase differ by ∼77° with respect to the dihedral angle defined by C3–C4–C4′–N. Other differences in the ketimine conformations seen in ColD and AATase include the positioning of the α-carboxylate moiety. In AATase, it participates in a salt bridge with Arg 386, whereas in ColD, it lies close to His 188.
Figure 6.
Comparison of the ColD glutamate ketimine intermediate with that observed in aspartate aminotransferase. Those residues highlighted in blue bonds correspond to the ColD model, whereas those amino acids depicted in green bonds correspond to aspartate aminotransferase (PDB accession no. 1MAQ). Residue labels beginning with an asterisk refer to aspartate aminotransferase.
It is important to note that the orientation of the glutamate ketimine intermediate observed in AATase (Fig. 6) can be easily accommodated in the ColD active site if the side chain of Trp 88 moves slightly. In this orientation the α-carboxylate of the ketimine intermediate in ColD would participate in a salt bridge with the guanidinium group of Arg 331 and the side chain carboxylate would be situated near the imidazole group of His 215. Importantly, this orientation of the ketimine in the ColD active site places the side chain of His 188 on the re face of its sp2 hybridized α-carbon. As will be discussed below, it has been postulated that bond cleavage at the sugar C3′ and bond formation at the cofactor C4′ most likely occurs on the re face of the Schiff base complex (Alam et al. 2004). The unusual binding of the glutamate ketamine observed in the ColD active site, where the planarity of the intermediate is broken, may simply be a function of the pH (6.0) at which the crystals were obtained.
While GDP was included in the initial crystallization trials, it was not observed in any of the electron density maps. Repeated attempts at soaking crystals in solutions containing GDP up to 10 mM were also unsuccessful. From the structure of the ColD dimer it is clear that a rather large open cleft extends from the ketimine intermediate to the surface. Preliminary attempts at collecting X-ray data from crystals grown in the presence of GDP-4-keto-6-deoxymannose have, however, met with some limited success (unpublished data). In a map calculated with coefficients of the form (Fo − Fc), a tube of electron density was observed that extends from the PLP cofactor toward the protein surface. Embedded in this density were two large peaks (4σ) separated by ∼2.8 Å. Assuming that these two peaks correspond to the phosphoryl moieties of the substrate, it was possible to build a model for it with its sugar C4′ keto group placed near the PLP cofactor and the phosphoryl and GDP-ribosyl groups extending toward the solvent. Importantly, this model places the phosphoryl moieties near the positively charged side chains of Arg 331 in subunit 1 and Arg 219 and Arg 250 in subunit 2. If this model is correct, most of the ligand/protein binding interactions would be primarily through the sugar and the phosphoryl groups of the substrate. It is not known whether ColD exhibits a strict specificity for GDP or can utilize other nucleotide-linked sugars.
ColD functions in the GDP–colitose biosynthetic pathway by removing the hydroxyl group at C3′. Like colitose, tyvelose is another 3,6-dideoxyhexose that occurs in the O-antigens of some Gram-negative bacteria. However, the removal of its 3′-hydroxyl group occurs via a completely different reaction mechanism. Specifically, the dehydration at C3′ is catalyzed by the E1/E3 complex, which requires NAD, FAD, and [2Fe-2S] clusters (He and Liu 2002). Indeed, ColD is an especially intriguing sugar dehydratase in that it lacks any requirement for NAD, FAD, and iron–sulfur centers.
A proposed mechanism for ColD has been previously put forth (Alam et al. 2004), and is presented in adapted forms in Schemes 2 and 3. In the first step (Scheme 2), PLP is converted to PMP through its reaction with l-glutamate. A catalytic base is required for this process which, based on the structure presented here, we propose to be His 188. This residue resides within ∼4.4 Å of the glutamate ketimine α-carbon. If glutamate is modeled into the active site on the basis of the observed ketimine orientation in AATase, His 188 is positioned such that its imidazole ring is ∼3 Å from the α-carbon proton.
Scheme 3.
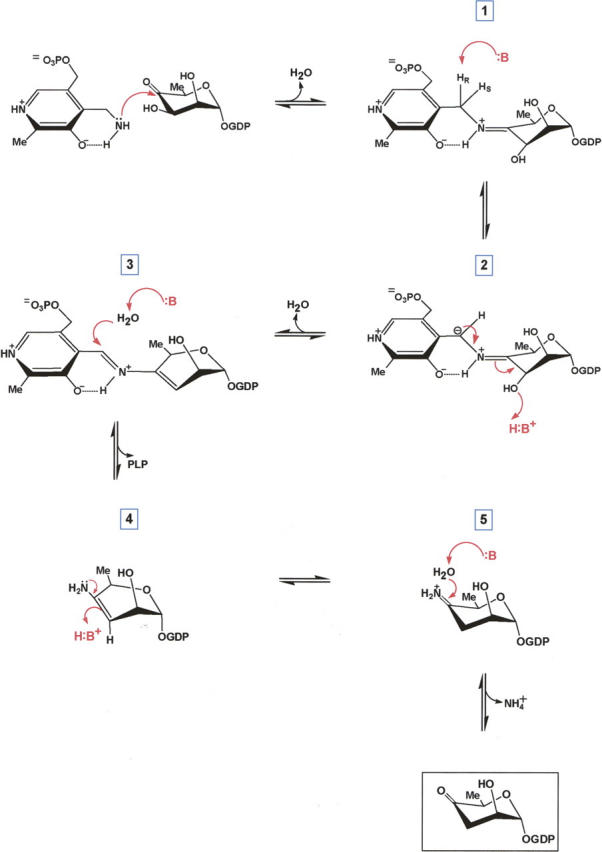
Following the production of PMP, a Schiff base is formed between the C4′ sugar carbon and the cofactor, labeled 1 in Scheme 3. From detailed biochemical investigations, it is known that deprotonation at C4′ of the cofactor is stereospecific, and that a catalytic base removes the pro-R hydrogen (Alam et al. 2004). Again, based on the ColD model presented here, it can be speculated that His 188 serves as the catalytic base to initiate the deprotonation event leading to the intermediate labeled 2 in Scheme 3. The ColD active site is markedly devoid of potential bases other than His 188, His 215, Asp 159, and Glu 162. As previously noted, both Asp 159 and Glu 162 are involved in hydrogen bonding interactions with the cofactor ring, and most likely are not involved in catalysis. Likewise, His 215 is not in the correct position to promote catalysis in light of previously reported stereochemical considerations (Alam et al. 2004).
The expulsion of the sugar C3′ hydroxyl group, as indicated in 2, requires an active site acid. Most likely His 188, which is now protonated, functions in this capacity. Following the departure of the C3′ hydroxyl moiety in the form of water, a Δ3,4-aminomannoseen intermediate is formed as shown in 3. Hydrolysis of this intermediate leads to the loss of PLP and the intermediate shown in 4 of Scheme 3. We propose that His 188 serves to activate a water molecule for this hydrolysis event. It has been demonstrated that in the next step labeled 4 in Scheme 3, the incorporation of a hydrogen at the sugar C3′ is stereospecific (Alam et al. 2004). Again, His 188 probably functions in this step as the active site acid. It is presently not known whether the final expulsion of ammonia, as indicted in 5 in Scheme 3, occurs on the enzyme or in solution.
Why did ColD evolve with a histidine in place of the lysine that normally forms an internal aldimine with PLP? We hypothesize that this occurred because of the unique chemistry involved in the reaction. In going from 3 to 4 in Scheme 3, an enzymatic base is required. In theory, this could be accomplished by a lysine residue. However, in this case, the lysine would most likely react directly with the C4′ of the cofactor to form an internal aldimine. As such, the next step leading to 4, which is the stereospecific protonation at C3′, would never occur given the arrangement of side chains in the active site of ColD. By having a histidine residue in place of a lysine, the formation of an internal aldimine is prevented, and importantly, histidine provides the functional group necessary for the reaction to proceed from 3 to 4. Experiments designed to test this hypothesis are presently underway.
From a chemical viewpoint, the in vitro synthesis of nucleotide-linked sugars is a complicated and arduous process at best. But nature has developed a large repertoire of enzymes for the specific and efficient biosynthesis of unique carbohydrates. To better understand the determinants of protein-substrate specificities and the fundamental reaction mechanisms utilized in these biosynthetic pathways, the structures and functions of the enzymes must be carefully studied. The models of ColD presented here provide new and powerful structural foundations upon which to more fully explore this fascinating PLP-dependent enzyme.
Materials and methods
Molecular cloning of the ColD gene
Genomic DNA from enterohemorrhagic E. coli O55:H7 was provided by the laboratory of Dr. Rodney Welch, Department of Medical Microbiology and Immunology, University of Wisconsin–Madison. The colD gene was PCR-amplified using primers that introduced a 5′ NdeI site and a 3′ XhoI site. The purified PCR product was A-tailed and ligated into the pGEM-T (Promega) vector for screening and sequencing. A ColD-pGEM-T vector construct of the correct sequence was then appropriately digested and ligated into a pET28b(+) vector that had been modified to provide an N-terminal TEV protease recognition site (Thoden et al. 2005).
DH5-α E. coli cells were transformed with the resulting plasmid and plated onto LB Agar Kan plates. Multiple colonies were picked to grow overnight in LB Kan medium from which plasmid DNA was isolated. Plasmids were tested for the presence of the colD gene by digestion with NdeI and XhoI.
Protein expression and purification
The ColD-pET28JT plasmid was used to transform Rosetta (DE3) E. coli cells (Novagen). The culture was grown at 37°C with shaking until an optical density of 0.9 was reached at 600 nm. The flasks were then cooled on ice and induced with 1 mM IPTG. Cells were allowed to express at 16°C for 24 h after induction with IPTG.
The cells were harvested and disrupted by sonication on ice. The lysate was cleared by centrifugation and ColD was purified utilizing Ni-NTA resin (Qiagen) according to the manufacturer's instructions. The N-terminal His-tag was cleaved from the protein by the addition of both 1 mM DTT and TEV protease in a 1:50 (TEV protease:ColD) molar ratio. The cleavage reaction was carried out at 4°C for 36 h and subsequently passed through the nickel column to remove the TEV protease. The ColD protein sample was dialyzed against 25 mM Tris-HCl and 200 mM NaCl (pH 8.0) with the addition of 1 mM PLP and 2 mM glutamate or α-ketoglutarate. Following dialysis, the sample was concentrated to 20 mg/mL based on an extinction coefficient of 0.95 (mg/mL)−1 · cm−1 as calculated with the ProtParam tool on the ExPASy Proteomics Server.
Crystallization of ColD
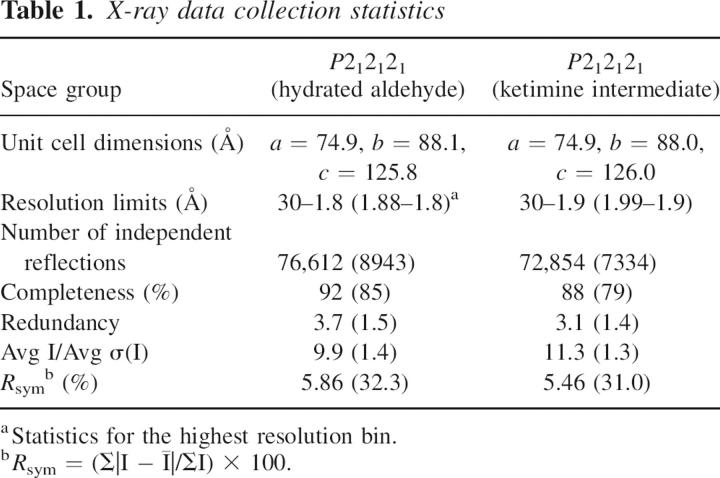
Crystallization conditions were first surveyed by the hanging drop method of vapor diffusion with a sparse matrix screen developed in the laboratory. Large crystals were subsequently grown via batch methods by mixing 20 μL of a protein solution with 20 μL of precipitant. Protein concentrations were typically at 20 mg/mL and the solutions contained 25 mM Tris (pH 8.0), 200 mM NaCl, 1 mM PLP, and 2 mM glutamate or α-ketoglutarate. The precipitant solutions contained 28% poly(ethylene glycol) 3400, 100 mM MES (pH 6.0), 400 mM MgCl2, 1 mM PLP, 2 mM glutamate or α-ketoglutarate, and 1 mM GDP. The crystals grew to maximum dimensions of 1.8 × 0.5 × 0.5 mm in ∼2 d. They belonged to the space group P212121, with unit cell dimensions of a = 74.9, b = 88.1, and c = 125.8, and contained a dimer in the asymmetric unit.
Structural analysis of ColD
X-ray data sets were collected from two different crystal complexes. In one form the protein contained hydrated PLP in its active site, while in the second form the protein contained the trapped glutamate ketimine intermediate. All X-ray data were measured with a Bruker HISTAR area detector system. The X-ray source was CuKα radiation from a Rigaku RU200 X-ray generator operated at 50 kV and 90 mA. The X-ray data were processed with XDS (Kabsch 1993) and internally scaled with XSCALIBRE (I. Rayment and G. Wesenberg, unpubl.). Relevant X-ray data collection statistics are presented in Table 1.
Table 1.
X-ray data collection statistics
The structure of the ColD/hydrated PLP complex was solved via molecular replacement with the program Phaser and employing ArnB, a 4-amino-4-deoxy-l-arabinose lipopolysaccharide-modifying enzyme (PDB accession no. 1MDO) as a search model (Noland et al. 2002). The electron densities corresponding to the two subunits in the asymmetric unit were averaged with the software package DM (Cowtan and Main 1998), and a model was manually built on the basis of this averaged map. The model was then placed back into the unit cell for subsequent least-squares refinement with the software package TNT (Tronrud et al. 1987). Alternate cycles of manual model rebuilding and least-squares refinement reduced the R-factor to 17.0% at 1.8 Å resolution. The structure of the trapped glutamate ketimine intermediate was solved via difference Fourier methods. Alternate cycles of manual model rebuilding and least-squares refinement of this model reduced the R-factor to 17.6% at 1.9 Å resolution. Relevant refinement statistics are given in Table 2.
Table 2.
Least-squares refinement statistics
In each subunit (for both complexes) Met 193, Val 334, and Asn 354 adopt ϕ, ψ angles that are outside of the allowed regions of the Ramachandran plot (all near the “nucleophile elbow” region of ϕ = ∼60°, ψ = ∼−100°). The electron densities for these residues are unambiguous. Met 193 is positioned ∼10 Å from the phosphate of PLP, and is located in a sharp, nonclassical reverse turn. The side chain of Val 334 lies within ∼5 Å of the PLP, and is situated in a Type II′ turn. Asn 354 sits in a surface loop ∼20 Å from the active site. Other than these amino acids, 89.3%, 10.5%, and 0.1% of the remaining residues fall into the “most-favored,” “additionally allowed,” and “generously allowed” regions of the Ramachandran plot, respectively. Both complexes contain one magnesium ion. This cation resides near a crystalline lattice interface. While 1 mM GDP was included in the crystallizations, it was not observed in the electron density maps.
PDB accession numbers
X-ray coordinates have been deposited in the Protein Data Bank with accession numbers 2GMS and 2GMU.
Acknowledgments
We thank Professors Ivan Rayment and W.W. Cleland for helpful discussions. We are indebted to Professor Rodney Welch for graciously providing genomic DNA. This research was supported by a grant from the NIH (DK47814 to H.M.H.).
Footnotes
Reprint requests to: Hazel M. Holden, 433 Babcock Drive, Department of Biochemistry, University of Wisconsin––Madison Madison, WI 53706, USA; e-mail: Hazel_Holden@biochem.wisc.edu; fax: (608) 262-1319.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062328306.
References
- Alam J., Beyer N., Liu H.W. 2004. Biosynthesis of colitose: Expression, purification, and mechanistic characterization of GDP-4-keto-6-deoxy-d-mannose-3-dehydrase (ColD) and GDP-l-colitose synthase (ColC). Biochemistry 43: 16450–16460. [DOI] [PubMed] [Google Scholar]
- Bastin D.A. and Reeves P.R. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 164: 17–23. [DOI] [PubMed] [Google Scholar]
- Beyer N., Alam J., Hallis T.M., Guo Z., Liu H.W. 2003. The biosynthesis of GDP-l-colitose: C-3 deoxygenation is catalyzed by a unique coenzyme B6-dependent enzyme. J. Am. Chem. Soc. 125: 5584–5585. [DOI] [PubMed] [Google Scholar]
- Cowtan K. and Main P. 1998. Miscellaneous algorithms for density modification. Acta Crystallogr Sect D 54: 487–493. [DOI] [PubMed] [Google Scholar]
- Eads J.C., Beeby M., Scapin G., Yu T.W., Floss H.G. 1999. Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase. Biochemistry 38: 9840–9849. [DOI] [PubMed] [Google Scholar]
- Edstrom R.D. and Heath E.C. 1965. Isolation of colitose-containing oligosaccharides from the cell wall lipopolysaccharide of Escherichia coli. Biochem. Biophys. Res. Commun. 21: 638–643. [DOI] [PubMed] [Google Scholar]
- Ghisalba O. and Nuesch J. 1981. IV. Identification of 3-amino-5-hydroxybenzoic acid as a direct precursor of the seven-carbon amino starter-unit. J. Antibiot. (Tokyo). Nocardia mediterranei. 34: 64–71. [DOI] [PubMed] [Google Scholar]
- Harris C.M., Johnson R.J., Metzler D.E. 1976. Band-shape analysis and resolution of electronic spectra of pyridoxal phosphate and other 3-hydroxypyridine-4-aldehydes. Biochim. Biophys. Acta 421: 181–194. [DOI] [PubMed] [Google Scholar]
- He X.M. and Liu H.W. 2002. Formation of unusual sugars: Mechanistic studies and biosynthetic applications. Annu. Rev. Biochem. 71: 701–754. [DOI] [PubMed] [Google Scholar]
- Hisatsune K., Kondo S., Isshiki Y., Iguchi T., Kawamata Y., Shimada T. 1993. O-antigenic lipopolysaccharide of Vibrio cholerae O139 Bengal, a new epidemic strain for recent cholera in the Indian subcontinent. Biochem. Biophys. Res. Commun. 196: 1309–1315. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander C. 1996. Mapping the protein universe. Science 273: 595–603. [DOI] [PubMed] [Google Scholar]
- Jansonius J.N. 1998. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 8: 759–769. [DOI] [PubMed] [Google Scholar]
- Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26: 795–800. [Google Scholar]
- Kennedy J.F. and White C.A. In Bioactive carbohydrates in chemistry, biochemistry, and biology. . 1983. Ellis Horwood Publishers, Chichester, UK.
- Kotandrova N.A., Gorshkova R.P., Zubkov V.A., Ovodov Iu S. 1989. The structure of the O-specific polysaccharide chain of the lipopolysaccharide of Yersinia pseudotuberculosis serovar VII. Bioorg. Khim. 15: 104–110. [PubMed] [Google Scholar]
- Lerouge I. and Vanderleyden J. 2002. O-antigen structural variation: Mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26: 17–47. [DOI] [PubMed] [Google Scholar]
- Luderitz O., Staub A.M., Stirm S., Westphal O. 1958. Colitose, 3-desoxy-l-fucose, a new sugar building block with immunospecific functions in the endotoxins, lipopolysaccharides, of some Gram-negative bacteria. Biochem. Z. 330: 193–197. [PubMed] [Google Scholar]
- Malashkevich V.N., Toney M.D., Jansonius J.N. 1993. Crystal structures of true enzymatic reaction intermediates: Aspartate and glutamate ketimines in aspartate aminotransferase. Biochemistry 32: 13451–13462. [DOI] [PubMed] [Google Scholar]
- Muldoon J., Perepelov A.V., Shashkov A.S., Gorshkova R.P., Nazarenko E.L., Zubkov V.A., Ivanova E.P., Knirel Y.A., Savage A.V. 2001. Structure of a colitose-containing O-specific polysaccharide of the marine bacterium Pseudoalteromonas tetraodonis IAM 14160(T). Carbohydr. Res. 333: 41–46. [DOI] [PubMed] [Google Scholar]
- Nishino J., Hayashi H., Ishii S., Kagamiyama H. 1997. An anomalous side reaction of the Lys303 mutant aromatic L-amino acid decarboxylase unravels the role of the residue in catalysis. J. Biochem. 121: 604–611. [DOI] [PubMed] [Google Scholar]
- Noland B.W., Newman J.M., Hendle J., Badger J., Christopher J.A., Tresser J., Buchanan M.D., Wright T.A., Rutter M.E., Sanderson W.E. et al. 2002. Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-l-arabinose lipopolysaccharide-modifying enzyme. Structure 10: 1569–1580. [DOI] [PubMed] [Google Scholar]
- Rupprath C., Schumacher T., Elling L. 2005. Nucleotide deoxysugars: Essential tools for the glycosylation engineering of novel bioactive compounds. Curr. Med. Chem. 12: 1637–1675. [DOI] [PubMed] [Google Scholar]
- Silipo A., Molinaro A., Nazarenko E.L., Gorshkova R.P., Ivanova E.P., Lanzetta R., Parrilli M. 2005. The O-chain structure from the LPS of marine halophilic bacterium Pseudoalteromonas carrageenovora-type strain IAM 12662T. Carbohydr. Res. 340: 2693–2697. [DOI] [PubMed] [Google Scholar]
- Thoden J.B., Timson D.J., Reece R.J., Holden H.M. 2005. Molecular structure of human galactokinase: Implications for Type II galactosemia. J. Biol. Chem. 280: 9662–9670. [DOI] [PubMed] [Google Scholar]
- Toney M.D. 2001. Computational studies on nonenzymatic and enzymatic pyridoxal phosphate catalyzed decarboxylations of 2-aminoisobutyrate. Biochemistry 40: 1378–1384. [DOI] [PubMed] [Google Scholar]
- Toney M.D. 2005. Reaction specificity in pyridoxal phosphate enzymes. Arch. Biochem. Biophys. 433: 279–287. [DOI] [PubMed] [Google Scholar]
- Tramonti A., John R.A., Bossa F., De Biase D. 2002. Contribution of Lys276 to the conformational flexibility of the active site of glutamate decarboxylase from Escherichia coli. Eur. J. Biochem. 269: 4913–4920. [DOI] [PubMed] [Google Scholar]
- Tronrud D.E., Ten Eyck L.F., Matthews B.W. 1987. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallogr A 43: 489–501. [Google Scholar]
- Weymouth-Wilson A.C. 1997. The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 14: 99–110. [DOI] [PubMed] [Google Scholar]
- Xiang S.H., Haase A.M., Reeves P.R. 1993. Variation of the rfb gene clusters in Salmonella enterica. J. Bacteriol. 175: 4877–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-J. and Matthews B.W. 1995. EDPDB: A multifunctional tool for protein structure analysis. J. Appl. Crystallogr. 28: 624–630. [Google Scholar]