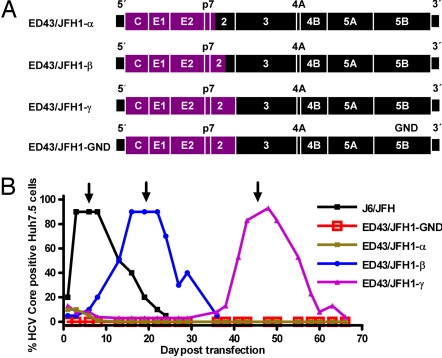
Fig. 1.
ED43/JFH1 recombinants and their viability in Huh7.5 cells. (A) Genome map of cDNA clones (ED43, purple; JFH1, black). ED43/JFH1-α had its 3′-terminal intergenotypic junction at nt 2,866/2,867 (H77 reference (AF009606) nt 2,867/2,868), whereas the junction of ED43/JFH1-β was at nt 3,185/3,186 (H77 reference nt 3,186/3,187). The complete ED43 NS2 gene was included in ED43/JFH1-γ and the replication-deficient control ED43/JFH1-GND. (B) Huh7.5 cells were transfected in parallel with RNA transcripts from pJ6/JFH, pED43/JFH1-α, -β, -γ, and -GND. After immunostaining, the percentage of HCV Core-positive cells was scored by using confocal fluorescence microscopy. One milliliter of supernatant from day 6 (J6/JFH; ≈104.4 TCID50), day 19 (ED43/JFH1-β; ≈102.9 TCID50 and ED43/JFH1-GND), or day 45 (ED43/JFH1-γ; ≈102.9 TCID50) was used for first passage (arrows) (SI Fig. 6). ED43/JFH1-α Core-positive cells were detected until day 19 but not thereafter.