Fig. 4.
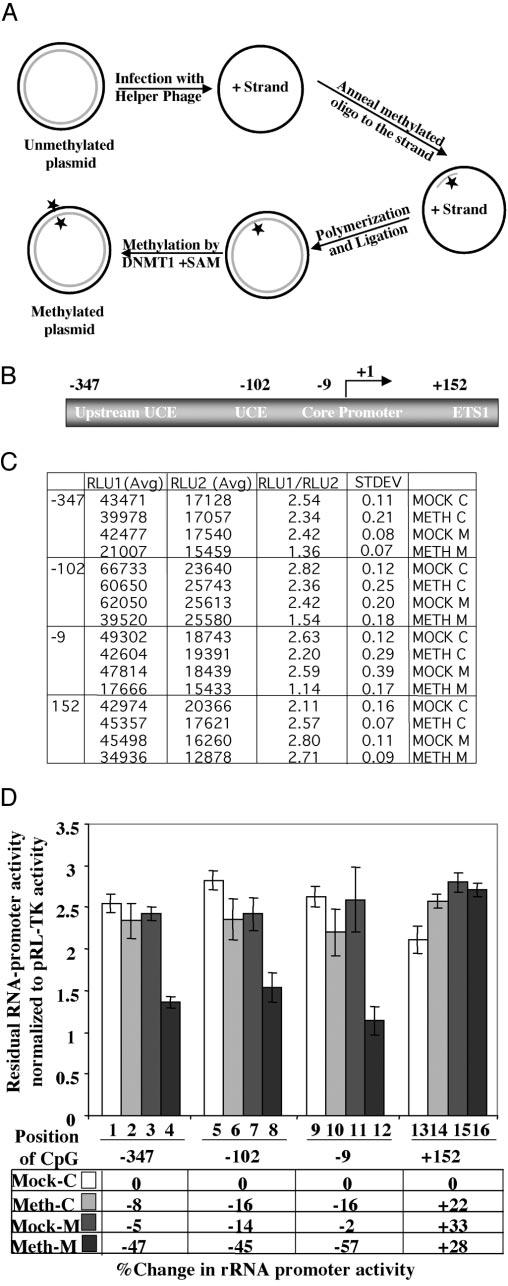
A, schematic depiction of site-specific methylation technique. B, location of CpGs on human rRNA gene subjected to site-specific methylation to study their effect on the promoter activity. These sites were selected because of the ability to generate primers spanning these regions. The construction of pHrD-IRES-Luc methylated at specific sites is described under “Materials and Methods.” C and D, plasmids (500 ng) methylated in both strands (Meth M), methylated in the (−)-strand (Mock M), or nonmethylated controls (Mock C and Meth C) along with pRL-TK (50 ng) were transfected into 2 × 105 HepG2 cells, and both firefly (RLU1) and Renilla (RLU2) luciferase activities were measured 24 h post-transfection using the Dual Luciferase Assay kit. The data of a representative experiment are tabulated in C and graphically represented in D.
