Fig. 7. Analysis of methyl-binding protein association with methylated/unmethylated human rRNA promoter by ChIP analysis.
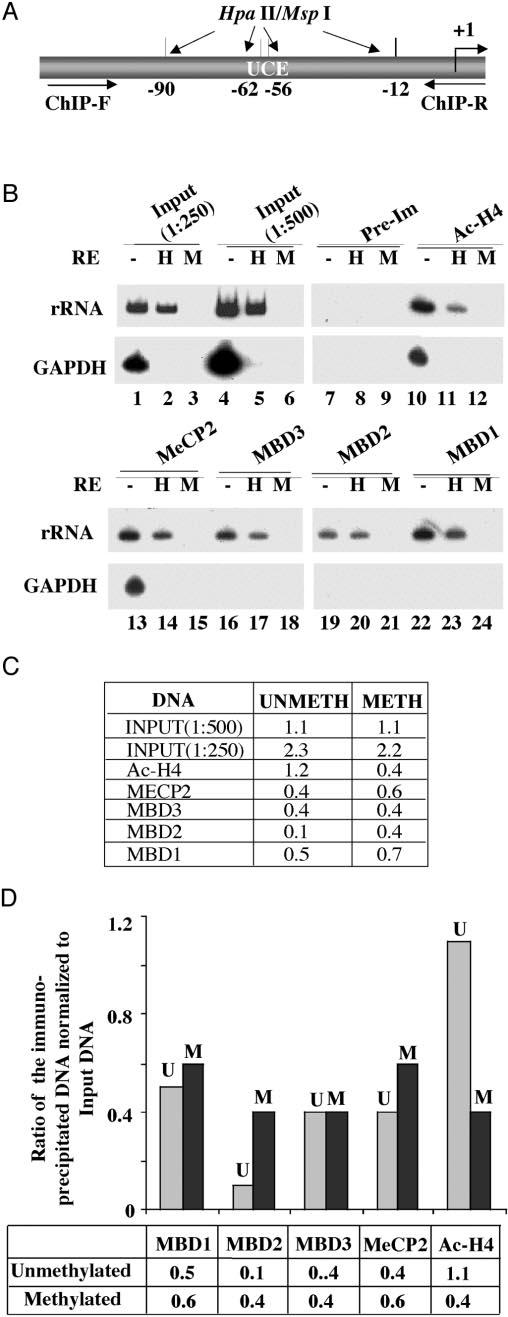
A, schematic presentation of the rRNA promoter region amplified and the locations of HpaII sites. B, formaldehyde cross-linked chromatin was precleared and immunoprecipitated overnight with antisera specific for MBDs, acetylated histone H3, or preimmune sera. The immune complexes were pulled down by protein A/G beads, washed with different buffers, eluted, and de-cross-linked. DNAs pulled down by different antibodies as well as input DNA were divided into three identical fractions that were either mock-digested or digested with HpaII (H) or MspI (M). An aliquot of the product was subjected to semiquantitative PCR with 32P-labeled forward primer specific for rRNA or GAPDH promoters. The reaction products were separated on polyacrylamide (8% for rRNA and 6% for GAPDH), and the dried gel was subjected to autoradiography (2-h exposure for rRNA and overnight exposure for GAPDH) and PhosphorImager analysis. C and D, quantitative analysis and graphical representation of the association of different MBDs with rDNA promoter. The Volume Analysis program of the ImageQuant software was used to quantify the 32P signal. Association with methylated promoter = HpaII signal in ChIP DNA/HpaII signal in input (1:500 dilution). Association with the unmethylated promoter = signal in undigested minus signal in HpaII-digested ChIP DNA/input signal in undigested minus HpaII-digested DNA (1:500 dilution). U and M, methylated and unmethylated rRNA promoters, respectively.
