Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Na2SO4-soaking form | NaCl/Tris form | |
|---|---|---|
| Data collection | ||
| Crystal-to-detector distance (mm) | 155 | 155 |
| Wavelength (Å) | 0.8 | 1.0 |
| Exposure time (s) | 1 | 3 |
| Total rotation range (°) | 180 | 180 |
| Space group | P212121 | P212121 |
| Unit-cell parameters | ||
| a (Å) | 49.88 | 46.13 |
| b (Å) | 57.85 | 57.29 |
| c (Å) | 74.33 | 72.89 |
| Resolution limits (Å) | 50–1.3 (1.40–1.30) | 30–1.5 (1.55–1.50) |
| Mosaicity | 0.45 | 1.13 |
| Total No. of reflections | 331403 | 195477 |
| Unique reflections | 52552 | 31471 |
| Redundancy | 6.3 (3.4) | 6.2 (2.6) |
| Completeness (%) | 96.1 (87.5) | 98.8 (89.8) |
| I/σ(I) | 14.1 (1.6) | 23.6 (1.5) |
| Rmerge† (%) | 9.0 (33.4) | 5.4 (40.4) |
| Refinement | ||
| Resolution limits (Å) | 50–1.3 (1.31–1.30) | 30–1.5 (1.52–1.50) |
| Reflections in test set (%) | 5 | 5 |
| R‡ (%) | 21.1 (27.4) | 22.3 (30.0) |
| Rfree§ (%) | 22.6 (29.4) | 24.6 (30.8) |
| No. of protein atoms | 1845 | 1845 |
| No. of ion atoms | 10¶ | 9†† |
| No. of water molecules | 292 | 236 |
| R.m.s.d. bond lengths (Å) | 0.013 | 0.007 |
| R.m.s.d. bond angles (°) | 1.6 | 1.8 |
| PDB code | 2de9 | 2de8 |
R
merge = 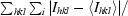
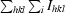 .
.
R = 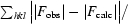
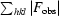 .
.
R free is equivalent to R, but is calculated using a test set of reflections excluded from the final refinement stages.
Two sulfates.
One Cl− and one tris(hydroxymethyl)aminomethane molecule.
