FIGURE 1. DNMT1 and -3B synergistically maintain rDNA promoter methylation.
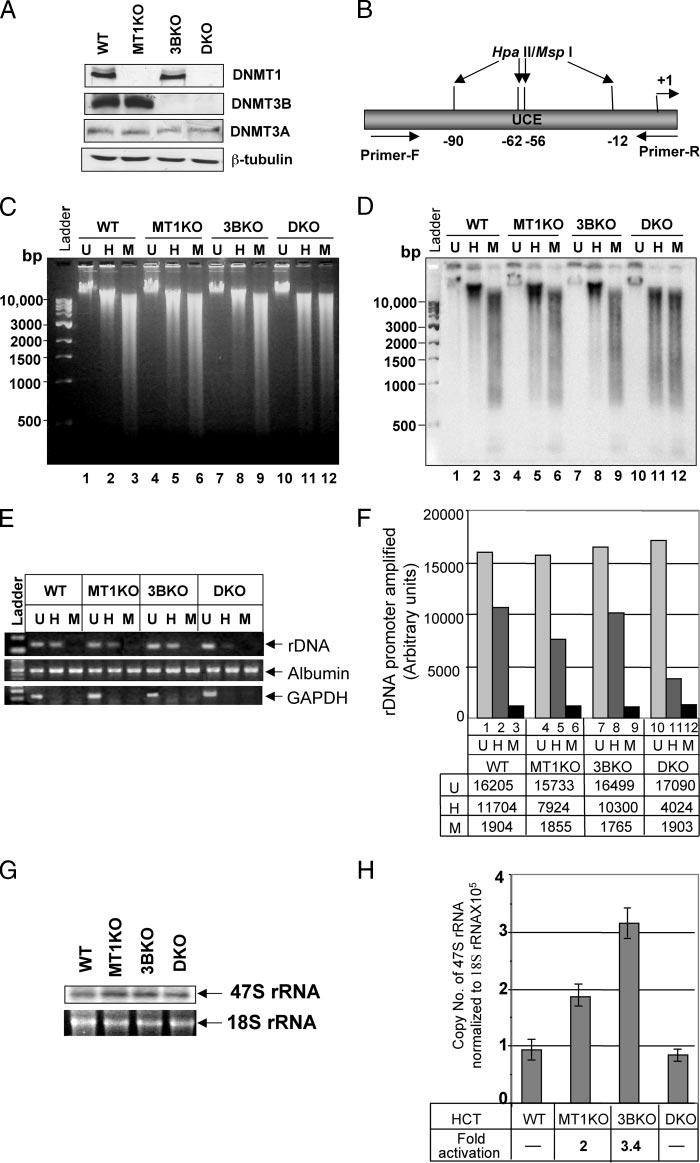
A, whole cell extracts from HCT116 (WT ), DNMT1−/− (MT1KO), DNMT3B−/− (3BKO), and DNMT1−/− /DNMT3B−/− (DKO) cells were separated by SDS-PAGE and Western blot analysis was performed with specific antibodies. B, schematic presentation of the rDNA promoter region used as probe for Southern blot analysis. C, genomic DNA from the cells were either mock-digested (U, Uncut), and digested with Hpall (H) or MspI (M). The digested products were separated on a 0.8% agarose gel and stained with ethidium bromide. D, DNA was transferred to nylon membrane and probed with a32P-labled rDNA promoter probe. E, rDNA, GAPDH, and albumin promoters were amplified from mock-digested (U, Uncut), Hpall (H), or MspI (M) digested genomic DNA from W T HCT116, MT1KO, 3BKO, and DKO cells and resolved on 1.5% agarose gel. F, quantitative analysis of the PCR amplified rDNA products. G, total RNA isolated from the cells was resolved on a 1.0% agarose/formaldehyde gel, transferred to nylon membrane, and probed with the 91-bp pre-RNA probe. H, total RNA from the cells was treated with RNase-free DNase I, reverse transcribed, and subjected to real time PCR using 47 S rRNA-specific primers. Data presented was normalized to 18 S rRNA. RNA without reverse transcription did not generate PCR product.
