FIGURE 2.
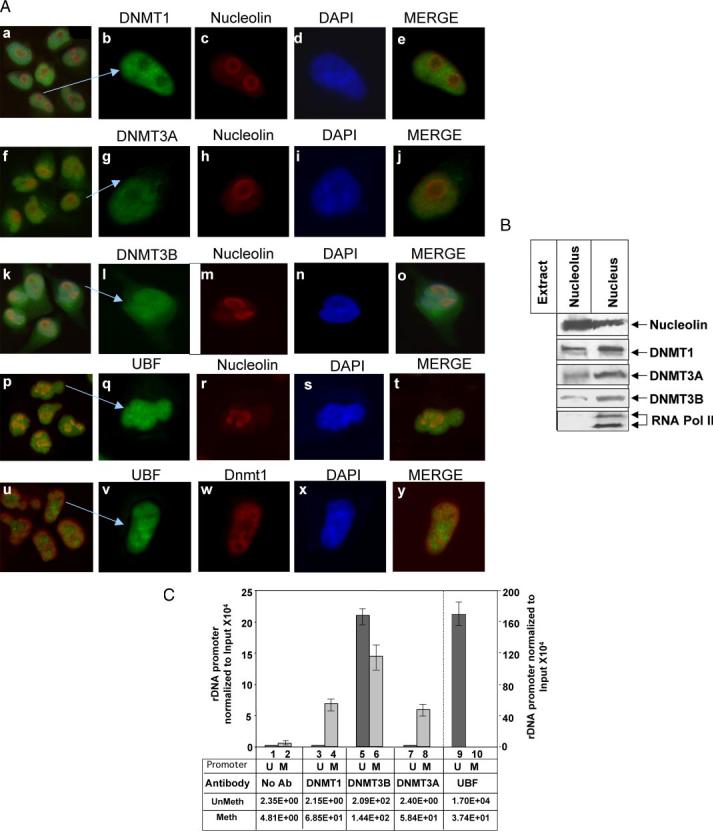
A, all three DNMTs localize in the nucleolus. HeLa cells were stained with TRITC-tagged mouse monoclonal antibody against nucleolin and with fluorescein isothiocyanate-tagged rabbit polyclonal antibody against DNMT1, 3A, -3B, or UBF (panels a–t). In a separate set, cells were stained with fluorescein isothiocyanate-tagged rabbit polyclonal antibody against UBF and TRITC-tagged mouse monoclonal antibody against DNMT1 (panels u–y). All five sets of cells were also stained with 4′,6-diamidino-2-phenylindole and visualized under a fluorescence microscope. B, all three DNMTs co-fractionated with nucleolin in the nucleolar fraction: nucleolar extract and nuclear extract (nucleolus and nucleoplasm) from HeLa cells (250 μg) were subjected to Western blot analysis with antibodies against nucleolin, RNA polymerase II, DNMT1, -3A, and -3B. C, DNMT1, -3B, and -3A are associated with methylated rDNA promoter. Formaldehyde cross-linked chromatin was precleared and immunoprecipitated overnight with antisera specific for DNMT1, DNMT3A, DNMT3B, UBF, or preimmune sera. The immune complexes were precipitated by protein A/G beads, washed with different buffers (detailed under “Experimental Procedures”), eluted, and un-cross-linked. DNAs pulled down by different antibodies as well as input DNA were divided into three identical fractions that were either mock digested or digested with HpaII or MspI. An aliquot of each digestion product was subjected to real time PCR with primers specific for rDNA promoter. Association of different DNMTs and UBF with the rDNA promoter was analyzed using a standard curve generated by serial dilution of the undigested input DNA. Association with methylated promoter equals HpaII signal in ChIP DNA/HpaII signal in input (1:300 dilution). Association with unmethylated promoter corresponds to the signal in undigested minus signal in HpaII-digested ChIP DNA/Input signal in undigested minus HpaII-digested DNA (1:300 dilution).U and M indicate methylated and unmethylated rDNA promoters, respectively.
