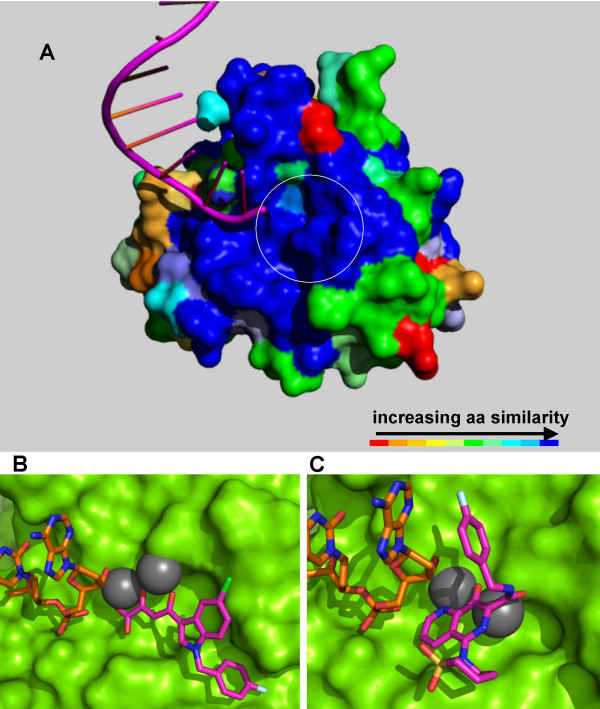
Figure 3.
Proposed binding mode of integrase strand transfer inhibitors (INSTIs) to FIV integrase. Panel A: A three-dimensional model of FIV-Pet IN catalytic core domain in complex with the transferred strand of viral DNA c. The enzyme is colored by sequence similarity with its HIV-1 orthologue [PDB:1BL3]. The level of similarity was calculated by the Swiss PDB Viewer (SPDBV) software. The color scale is that adopted by SPDBV. The transferred strand of proviral DNA is shown in magenta. Similarity is maximal at the level of the INSTI binding site. The INSTI binding site (indicated by a circle) is that calculated by some of us in previous works [16,20]. Panels B-C: Docking of CHI1019 (panel B) and L-870,810 (panel C) at the catalytic cavity of FIV IN. The protein is shown as Connolly surface (in green). Ligands are shown in CPK (carbon backbone in magenta). The terminal dinucleotide of 3' processed proviral DNA is shown in CPK (carbon backbone in orange). Metals are shown as spheres (in gray). Images prepared using Pymol (see Ref. [50]).