Abstract
PHO1 was previously identified in Arabidopsis (Arabidopsis thaliana) as a protein involved in loading inorganic phosphate (Pi) into the xylem of roots and its expression was associated with the vascular cylinder. Seven genes homologous to AtPHO1 (PpPHO1;1–PpPHO1;7) have been identified in the moss Physcomitrella patens. The corresponding proteins harbor an SPX tripartite domain in the N-terminal hydrophilic portion and an EXS domain in the conserved C-terminal hydrophobic portion, both common features of the plant PHO1 family. Northern-blot analysis showed distinct expression patterns for the PpPHO1 genes, both at the tissue level and in response to phosphate deficiency. Transgenic P. patens expressing the β-glucuronidase reporter gene under three different PpPHO1 promoters revealed distinct expression profiles in various tissues. Expression of PpPHO1;1 and PpPHO1;7 was specifically induced by Pi starvation. P. patens homologs to the Arabidopsis PHT1, DGD2, SQD1, and APS1 genes also responded to Pi deficiency by increased mRNA levels. Morphological changes associated with Pi deficiency included elongation of caulonemata with inhibition of the formation of side branches, resulting in colonies with greater diameter, but reduced mass compared to Pi-sufficient plants. Under Pi-deficient conditions, P. patens also increased the synthesis of ribonucleases and of an acid phosphatase, and increased the ratio of sulfolipids over phospholipids. These results indicate that P. patens and higher plants share some common strategies to adapt to Pi deficiency, although morphological changes are distinct, and that the PHO1 proteins are well conserved in bryophyte despite the lack of a developed vascular system.
Phosphorus (P) is an essential macronutrient for all living organisms. It serves a variety of basic biological functions as a structural element of many molecules, such as nucleic acids and phospholipids, and plays a pivotal role in energy metabolism, activation of metabolic intermediates, signal transduction cascades, and regulation of enzymes. Among the major nutrients required for plants, P is the most dilute and the least mobile in soil. Plants absorb P as orthophosphate (Pi, inorganic phosphate). As a consequence, plants have evolved a series of morphological, physiological, biochemical, and molecular adaptations leading to increased survival ability under limited Pi availability (Raghothama, 1999; Abel et al., 2002; Poirier and Bucher, 2002; Rausch and Bucher, 2002).
Pi limitation leads to morphological modifications, such as an increase in root-to-shoot ratio, increase in the length and density of root hairs, as well as of the proliferation of lateral roots (Raghothama, 1999; Bucher et al., 2001; Poirier and Bucher, 2002). These changes in root architecture markedly increase the root-soil interface, allowing the roots to explore a larger volume of soil. To solubilize Pi from their insoluble inorganic complexes and organic sources in soils, some plants increase the secretion of organic acids, such as malic and citric acids, from the root system. Plants have also been found to increase the production of nucleases and phosphatases to assist in the recovery of Pi from organic sources (Raghothama, 1999; Abel et al., 2002; Poirier and Bucher, 2002). Biochemical changes associated with Pi deficiency include remobilization of Pi associated with lipids by the replacement of phospholipids with galactolipids and sulfolipids (Essigmann et al., 1998).
At the molecular level, Pi transporters play an essential role in absorbing Pi from the environment and in translocating Pi within the plant. The PHT1 gene family in Arabidopsis (Arabidopsis thaliana) contains nine members and encodes high-affinity H+-Pi cotransporters (Muchhal et al., 1996; Smith et al., 1997; Mudge et al., 2002; Rausch and Bucher, 2002). Most of these genes, which are preferentially expressed in roots, demonstrate a specific response to Pi stress, including rapid up-regulation of expression but return to a basal expression level upon resupply of Pi (Karthikeyan et al., 2002; Mudge et al., 2002). Some PHT1 genes are expressed in plant tissues other than roots (e.g. stems, leaves, flowers), indicating that the corresponding proteins may have roles in Pi transport and homeostasis beyond Pi uptake from the soil (Karthikeyan et al., 2002; Mudge et al., 2002). Reverse genetics has shown that Arabidopsis Pht1;1 and Pht1;4 function in Pi acquisition from both low- and high-Pi environments (Misson et al., 2004; Shin et al., 2004).
The Arabidopsis PHO1 gene has been cloned by a map-based cloning strategy using a mutant showing severe deficiency in transferring Pi acquired by the root into the xylem vessel (Poirier et al., 1991; Hamburger et al., 2002). The N-terminal half of PHO1 is mainly hydrophilic and harbors a tripartite SPX domain, whereas the C-terminal half has at least six predicted transmembrane domains and harbors an EXS domain (Wang et al., 2004). The gene is predominantly expressed in the root and overexpressed during Pi starvation. AtPHO1 promoter-GUS reporter gene studies revealed predominant expression of the promoter in the stelar cells of the root and the lower part of the hypocotyl, which is consistent with the role of the AtPHO1 gene in loading Pi into the xylem of the roots (Hamburger et al., 2002).
Ten additional genes showing high homology with PHO1 are present in the Arabidopsis genome, forming a novel class of proteins potentially involved in Pi transport and homeostasis in plants. cDNAs of all 11 members of the PHO1 gene family have been cloned and sequenced (Wang et al., 2004). Analysis of the activity of the promoter of all PHO1 homologs using promoter-GUS fusions revealed predominant expression in the vascular tissues of roots, leaves, stems, or flowers. GUS expression was also detected for several promoters in nonvascular tissues, including hydathodes, trichomes, root tip, root cortical/epidermal cells, and pollen grains, indicating the potential role of the PHO1 family proteins not only in the transfer of Pi to the vascular xylem of various tissues, but also in the homeostasis or transport of Pi in other cells, such as pollen or root epidermal/cortical cells.
The moss Physcomitrella patens has been developed as a model system for plant biology (Reski, 1999; Cove, 2000; Schaefer, 2002; Frank et al., 2005; Cove et al., 2006). The dominance of the haploid gametophyte in its life cycle greatly facilitates genetic studies and its simple tissue organization permits single-cell analysis during development. P. patens is the only land plant thus far in which gene targeting occurs with an efficiency similar to that observed in yeast (Saccharomyces cerevisiae; Schaefer, 2002). Although a broad range of experiments on Arabidopsis and field crops have been undertaken, resulting in the identification of genes involved in plant nutrition, thus far the study of mineral nutrition and adaptive response to nutrient deficiency has not been well characterized in moss. This work describes the cloning and characterization of seven PHO1 homologs in P. patens, including their tissue expression profile and their regulation by Pi levels. Several morphological and biochemical changes induced by Pi deficiency in P. patens have also been characterized.
RESULTS
Cloning the P. patens PHO1 Gene Family
Analysis of the P. patens EST database (http://moss.nibb.ac.jp) using the TBLASTN program with the Arabidopsis PHO1 amino acid sequence revealed five cDNA clones (pdp09094, pdp00719, pdp32726, pdp36370, and pdp42910) encoding proteins showing homology to both the hydrophilic and hydrophobic regions of AtPHO1. These clones were acquired from the RIKEN BioResource Center of Japan and were sequenced. Sequencing results indicated that these clones represented three distinct genes, which were named PpPHO1;1, PpPHO1;2, and PpPHO1;3. Similar analysis of the P. patens genome sequence database (http://genome.jgi-psf.org/Physcomitrella) also revealed the presence of four potential additional genes encoding proteins with homology to AtPHO1. Isolation of cDNA clones for these four genes using reverse transcription (RT)-PCR confirmed that these genes were distinct and were expressed in P. patens protonemata. They were thus named PpPHO1;4 to PpPHO1;7. Coding sequences of all seven genes were deposited in GenBank.
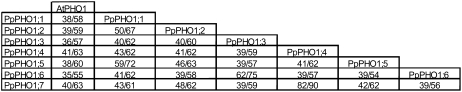
Pairwise comparison of the PpPHO1 protein family showed sequence conservation ranging from 39% amino acid identity and 54% similarity between PpPHO1;5 and PpPHO1;6 to 82% amino acid identity and 90% similarity between PpPHO1;4 and PpPHO1;7 (Fig. 1). This level of homology is similar to that observed between the various Arabidopsis PHO1 homologs (Wang et al., 2004). Comparison of the P. patens PHO1 proteins to the Arabidopsis PHO1 family revealed that the highest level of homology was between PpPHO1;4 and AtPHO1, with 41% amino acid identity and 63% similarity.
Figure 1.
Identity/similarity matrix for P. patens PHO1 family and Arabidopsis AtPHO1. The identity/similarity matrix was calculated using Matrix Global Alignment Tool software (http://bitincka.com/ledion/matgat). Amino acid identity and similarity are indicated by the first and second numbers, respectively.
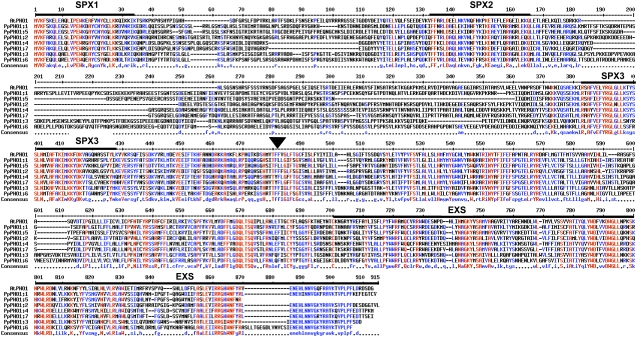
Alignment of the different P. patens PHO1 proteins with the Arabidopsis AtPHO1 revealed that the hydrophilic half of all PpPHO1 proteins harbors the SPX domain composed of three subdomains, which were separated by two areas of low similarity (Fig. 2). The entire hydrophobic half containing several potential transmembrane-spanning domains was well conserved among the PpPHO1 homologs and AtPHO1, including in the EXS domain (Fig. 2).
Figure 2.
Alignment of the P. patens PHO1 family and Arabidopsis AtPHO1. Alignment was done using the MultAlign program (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html). The locations of the three SPX subdomains and of the EXS domain are indicated by thick black bars over the sequence. The beginning of the hydrophobic C-terminal portion of the proteins is indicated by an arrowhead above the sequence.
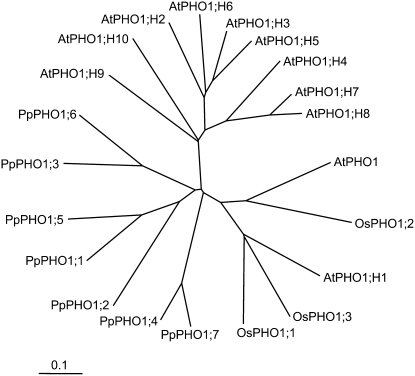
An unrooted phylogenetic tree constructed with Arabidopsis, P. patens, and rice (Oryza sativa) sequences revealed that most of the Arabidopsis PHO1 homologs form one cluster, with the exception of AtPHO1 and AtPHO1;H1, which form a second cluster and also includes the three rice OsPHO1 proteins, whereas the seven PpPHO1 members form a third group (Fig. 3).
Figure 3.
Unrooted phylogenetic tree of the PHO1 family in Arabidopsis, P. patens, and rice. The tree was made using the neighbor-joining method and displayed using the ClustalX program (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX). The three rice proteins OsPHO1;1, OsPHO1;2, and OsPHO1;3 correspond to the rice database entries Os01g0200, Os02g56510, and Os06g29790, respectively.
Expression Pattern of Members of the PpPHO1 Gene Family
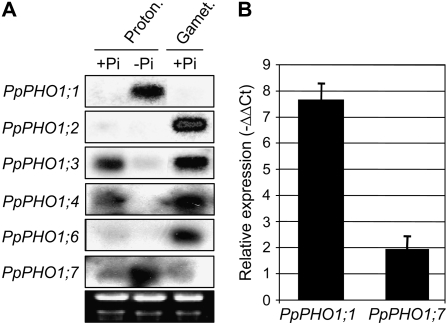
The expression profile of the PpPHO1 gene family was first investigated by northern blot using probes specific to each gene. In protonemata that were grown in a nutrient-rich medium containing 1.8 mm Pi, northern analysis showed robust expression of PpPHO1;3 and PpPHO1;4, weaker expression of PpPHO1;2, PpPHO1;6, and PpPHO1;7, whereas no expression was detectable for PpPHO1;1 and PpPHO1;5 (Fig. 4A; data not shown for PpPHO1;5). In the gametophore of plants grown in nutrient-rich medium, robust expression was detected for PpPHO1;2, PpPHO1;3, PpPHO1;4, and PpPHO1;6, weaker expression for PpPHO1;7, whereas no expression was detectable for PpPHO1;1 and PpPHO1;5 (Fig. 4A; data not shown for PpPHO1;5). Interestingly, the PpPHO1 genes showed different patterns of expression in protonemata grown in Pi-deficient medium. By comparing plants grown for 14 d in a medium containing either 1.8 mm Pi (+Pi) or without added Pi (−Pi), it was revealed that the expression of both PpPHO1;1 and PpPHO1;7 was up-regulated in protonemata under Pi-deficient conditions, whereas expression of PpPHO1;3 and PpPHO1;4 was down-regulated by Pi deficiency (Fig. 4A). Up-regulation of PpPHO1;1 and PpPHO1;7 expression in gametophores grown for 30 d in Pi-deficient medium was also observed by quantitative PCR analysis, with a stronger increase observed for PpPHO1;1 compared to PpPHO1;7 (Fig. 4B). RT-PCR analysis enabled the detection of transcripts for PpPHO1;1 and PpPHO1;5 in both protonemata and gametophores, as well as of PpPHO1;2 in protonemata, indicating a low level of transcription of these genes despite the failure to detect their expression by northern blot (data not shown).
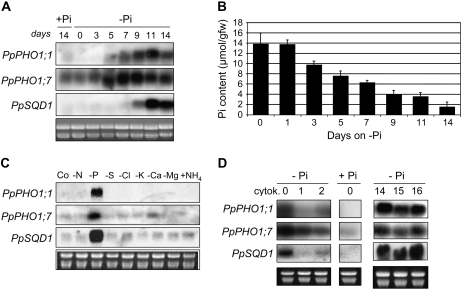
Figure 4.
Expression profile of members the PpPHO1 gene family. A, Northern blot was performed using gene-specific probes. Ethidium bromide staining of the gel is shown in the bottom image as loading controls. RNA was isolated either from protonemata grown on medium with (+Pi) or without (−Pi) Pi for 14 d or from gametophores with rhizoids grown on +Pi media. B, Expression of the PpPHO1;1 and PpPHO1;7 genes in gametophores grown in Pi-deficient medium relative to gametophores grown in Pi-sufficient medium, as measured by quantitative RT-PCR.
To further characterize the response of PpPHO1;1 and PpPHO1;7 to Pi deprivation, protonemata were first grown on +Pi medium and then transferred to −Pi medium for 3 to 14 d before being harvested for RNA extraction. The first increase in transcript of PpPHO1;1 detectable by northern blot occurred on day 5 and transcript abundance peaked on day 11, whereas for PpPHO1;7 the first detectable increase occurred on day 3 and peaked on days 7 to 9 (Fig. 5A). Analysis of Pi tissue content revealed a decrease of 33% and 48% of the Pi concentration in protonemata at 3 and 5 d following the shift to Pi-deficient medium, respectively (Fig. 5B).
Figure 5.
Modulation of gene expression by phosphate deficiency and cytokinin. A, Northern-blot analysis with RNA isolated from protonemata grown first for 6 d on medium with phosphate and then transferred to fresh medium with phosphate (+Pi) for 14 d, or medium without phosphate (−Pi) for 0 to 14 d. B, Pi content in protonemata grown as described in A. C, Northern-blot analysis using RNA isolated from protonemata grown for 14 d in complete medium (Co) or in medium deficient in either nitrogen (−N), phosphate (−P), sulfur (−S), chlorine (−Cl), potassium (−K), calcium (−Ca), magnesium (−Mg), or medium supplemented with ammonium (+NH4). D, Northern-blot analysis using RNA isolated from protonemata grown on medium with phosphate (+Pi) for 14 d (middle) or on medium without phosphate (−Pi) for 14 d and then transferred to −Pi medium supplemented with 3 μm 6-benzylaminopurine (cytokinin) for 1 and 2 d (left). As a control, protonemata grown on medium without phosphate (−Pi) for 14 d were transferred to fresh medium without Pi and without cytokinin for one and two additional days (days 15 and 16; right).
To determine whether deficiencies in other elements could affect the expression of the PpPHO1;1 and PpPHO1;7 genes, protonemata were grown for 14 d in medium deficient in various macro- and micronutrients and tissues were collected and analyzed by northern blot (Fig. 5C). Transcript of both PpPHO1;1 and PpPHO1;7 was strongly and specifically up-regulated by Pi deficiency.
Addition of cytokinin to Pi-deficient rice or Arabidopsis has been shown to attenuate expression of genes that are specifically up-regulated by Pi deficiency (Martin et al., 2000; Hou et al., 2005; Kobayashi et al., 2006; Lai et al., 2007). To reveal whether a similar mechanism exists in P. patens, protonemata grown for 14 d in Pi-deficient medium were transferred to Pi-deficient medium supplemented with the cytokinin 6-benzylaminopurine for up to 2 d (Fig. 5D). Expression of PpPHO1;1 was strongly suppressed by cytokinin from day 1, whereas repression of PpPHO1;7 by day 1 was weak but increased by day 2.
Expression Patterns of PpPHO1;1, PpPHO1;2, and PpPHO1;3 Revealed by Promoter-GUS Constructs
To further study the expression pattern of the first three PpPHO1 genes, a region upstream of the start codon of 1,173 bp for PpPHO1;1, 2,359 bp for PpPHO1;2, and 1,539 bp for PpPHO1;3 was used for promoter-GUS fusion constructs. Each construct was transformed into P. patens and the GUS expression profile of several independent clones was analyzed.
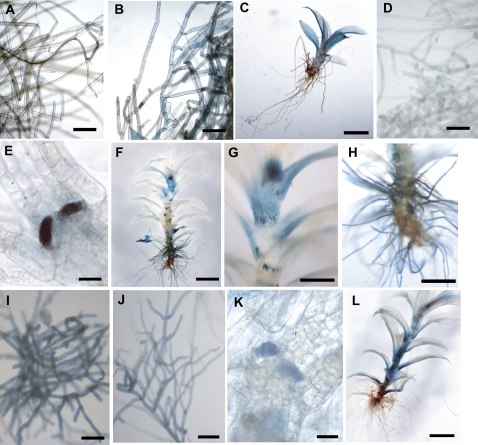
GUS expression from the PpPHO1;1, PpPHO1;2, and PpPHO1;3 promoters was observed in the protonemata, but in different intensity. Protonemata transformed with the PpPHO1;1∷GUS and PpPHO1;2∷GUS constructs were weakly stained after 24 h of incubation in substrate (Fig. 6, A and D), whereas those of PpPHO1;3 were strongly stained after 6 h (Fig. 6I). When grown on medium without Pi for 14 d, GUS expression from the PpPHO1;2 promoter was not observed in the protonemata even after 24 h of incubation in substrate (data not shown), whereas for PpPHO1;3 protonemata were stained after 6 h (Fig. 6J) but with lower intensity than those grown on normal Pi-containing medium. In contrast, the staining intensity observed from the PpPHO1;1 promoter in protonemata of Pi-deprived plants was stronger after 6 h of incubation compared to plants grown on Pi-rich medium and stained for 24 h (Fig. 6, A and B). GUS expression was not observed in buds but in the initiated leaves of young gametophores for both PpPHO1;2 and PpPHO1;3 promoters (Fig. 6, E and K), whereas no expression was observed in either tissues for PpPHO1;1. The gametophores displayed GUS expression for all three promoters, but in different intensities and patterns. The PpPHO1;1 promoter was weakly expressed across the blade of the majority of leaves (Fig. 6C). The PpPHO1;2 promoter was expressed strongly at the base of leaves, the apical portion of the gametophore, as well as in the auxiliary hairs that differentiate at the base of the leaves (Fig. 6, F and G). However, for PpPHO1;3, GUS expression was essentially localized at the base of the leaves (Fig. 6L). Finally, strong GUS expression was observed in rhizoids only with the PpPHO1;2 promoter (Fig. 6H).
Figure 6.
Localization of GUS activity in transgenic P. patens expressing PpPHO1;1, PpPHO1;2, and PpPHO1;3 promoter-GUS fusion constructs. A to C, PpPHO1;1 promoter activity in protonemata (A) and gametophore (C) for plants grown on +Pi medium and stained for 24 h and in protonemata of plants grown in −Pi medium and stained for 6 h (B). D to H, PpPHO1;2 promoter activity in protonemata (D) of plants grown on +Pi medium and stained for 24 h, and in emerging leaves (E), leafy gametophore (F and G), and rhizoids (H) for plants grown on +Pi medium and stained for 6 h. I to L, PpPHO1;3 promoter activity in protonemata of plants grown in +Pi medium (I) or −Pi medium (J) and stained for 6 h, as well as in emerging leaves (K) and leafy gametophores (L) of plants grown on +Pi medium and stained for 6 h. Bars = 100 μm (A, B, D, I, and J); 30 μm (E and K); 2 mm (C, F, and L); and 500 μm (G and H).
The Pi Deficiency Stress Response in P. patens
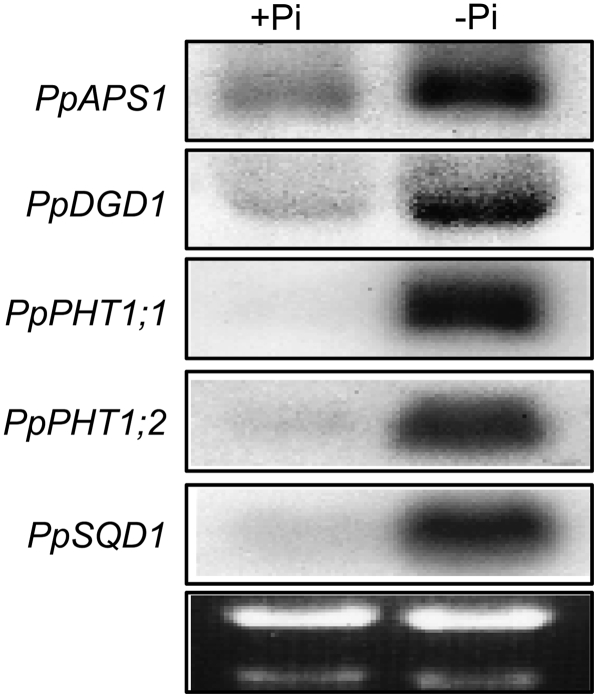
PpPHO1;1 and PpPHO1;7 are the first genes known from moss that are specifically up-regulated by Pi deficiency stress. It is known that, in Arabidopsis, as well as other higher plants, a number of genes are up-regulated in response to Pi starvation (Misson et al., 2005; Morcuende et al., 2007). To determine whether the genetic response of moss to Pi starvation is similar to that of Arabidopsis, regulation of several P. patens genes showing homology to Arabidopsis genes that are up-regulated in response to Pi deficiency was investigated. P. patens cDNAs encoding proteins showing homology to the following Arabidopsis proteins were used: cDNA pdp00526 showed homology to Arabidopsis APS1, encoding the small subunit of ADP-Glc pyrophosphorylase, a key enzyme of starch synthesis (Ciereszko et al., 2001), designated PpAPS1; cDNA pdp03201 showed homology to Arabidopsis DGD2, encoding UDP-Gal-dependent digalactosyldiacylglycerol synthase (Kelly and Dormann, 2002), designated PpDGD2; two cDNAs, pdp02478 and pdp33843, showed homology to Arabidopsis PHT1;4, encoding a high-affinity Pi transporter (Muchhal et al., 1996), designated PpPHT1;1 and PpPHT1;2, respectively; and cDNA pdp00870 showed homology to Arabidopsis SQD1, involved in the synthesis of sulfolipids (Essigmann et al., 1998), designated PpSQD1. Northern-blot analysis showed that all five genes were strongly up-regulated in protonemata grown in −Pi medium for 14 d compared to plants grown on +Pi medium (Fig. 7).
Figure 7.
Genes of P. patens up-regulated in protonemata of plants grown under Pi-deficient conditions. RNA was prepared from protonemata of plants grown on +Pi or −Pi medium for 14 d and analyzed by northern blot using gene-specific probes. Ethidium bromide staining of the gel is shown in the bottom image.
Time course of gene induction by Pi deficiency as well as specificity of the induction in plants grown in medium with different nutrient deficiencies was analyzed for PpSQD1. Clear up-regulation of PpSQD1 expression occurred on day 9 and peaked on day 11 after the shift to Pi-deficient medium (Fig. 5A). PpSQD1 was induced specifically by Pi deficiency and not by deficiencies in the other nutrients tested (Fig. 5C). Furthermore, addition of cytokinin to the growth medium strongly repressed expression of PpSQD1 in Pi-deficient plants (Fig. 5D).
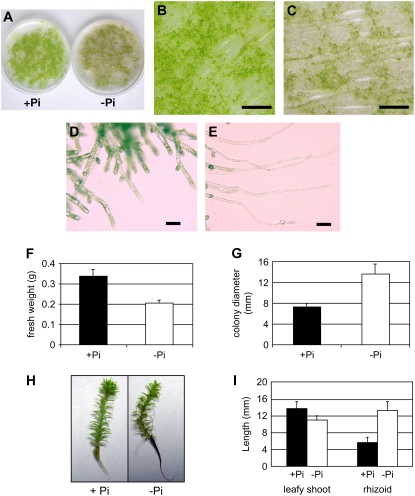
Morphological adaptation of P. patens grown under Pi-deficient conditions was investigated by growing plants for 14 to 90 d on +Pi or −Pi medium, conditions that lead to an 8- to 16-fold reduction in tissue Pi content (data not shown). Protonemata and gametophores grown on −Pi medium appeared brownish green (Fig. 8, A and C), whereas plants on +Pi medium were light green (Fig. 8, A and B). Gametophores produced on −Pi medium were less numerous than plants grown on +Pi medium (Fig. 8, B and C). In response to Pi deficiency, protonemata gave rise to a reduced number of chloronemata, but more caulonemata that were longer than for those grown on +Pi medium. In addition, Pi starvation inhibited the emergence of lateral branches from elongated caulonemata, whereas plants grown on +Pi medium branched regularly (Fig. 8, D and E). The reduction in side branches and the increased length of the caulonemata for plants grown under Pi-deficient conditions resulted in an increase in colony size by nearly 2-fold and a 38% decrease in biomass compared to plants grown on +Pi medium (Fig. 8, F and G). The leafy shoots of gametophores grown on −Pi medium for 90 d were slightly smaller, but had rhizoids approximately 2-fold longer compared to gametophores grown in +Pi medium (Fig. 8, H and I).
Figure 8.
Morphological alterations of P. patens grown under Pi-deficient conditions. A, P. patens grown on +Pi or −Pi medium for 28 d. B, Magnification of +Pi petri dish shown in A. C, Magnification of −Pi petri dish shown in A. D and E, Protonemata at the edge of colonies grown on +Pi (D) and −Pi (E) medium. F and G, Growth differences of P. patens colonies from protonemata distributed onto petri dishes containing +Pi or −Pi medium and grown for 14 d. The diameter of individual colonies (n = 20; F) and the plant biomass in each petri dish (n = 5; G) were measured. H and I, Gametophores grown in +Pi and −Pi medium for 90 d with measurements of the length of the leafy shoot and rhizoid (n = 8; I). Bars = 1 cm (B and C) and 60 μm (D and E).
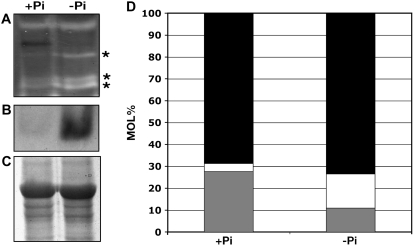
Extracts of soluble proteins from protonemata grown for 14 d in +Pi or −Pi medium were examined for the presence of proteins with ribonuclease activity and acid phosphatase activity by in-gel assays. Growth of protonemata under −Pi conditions resulted in the increased activity of three proteins with ribonuclease activity (Fig. 9A) and one protein with acid phosphatase activity (Fig. 9B). Analysis of lipids from protonemata grown for 14 d in +Pi or −Pi medium also revealed a decrease in the proportion of phospholipids and an increase in the proportion of galactolipids and sulfolipids (Fig. 9D).
Figure 9.
Changes in ribonuclease activity, acid phosphatase activity, and lipid composition in P. patens grown under Pi-deficient conditions. Protonemata were grown for 14 d in +Pi or −Pi medium, and extracts were analyzed for enzyme activity or lipid composition. A, Ribonuclease activity. Stars indicate locations of increased ribonuclease activity in −Pi sample. B, Acid phosphatase activity. C, Coomassie Blue staining of protein on SDS-PAGE as loading controls for A. D, Proportion, in mol%, of galactolipids (black), sulfolipids (white), and phospholipids (gray).
DISCUSSION
P. patens PHO1 proteins shared the main features found in all members of the Arabidopsis PHO1 family, namely, proteins with a large N-terminal hydrophilic domain containing the tripartite SPX domain and a hydrophobic C-terminal domain containing at least six transmembrane regions and the EXS domain (Wang et al., 2004). PHO1 thus represents a family of proteins not only conserved in a broad range of vascular plants, including angiosperms and gymnosperms, but also in nonvascular plants, such as the bryophyte P. patens.
The seven PpPHO1 genes displayed distinct expression profiles as revealed by northern-blot analysis and extended by analysis of promoter-GUS expression profiles for PpPHO1;1, PpPHO1;2, and PpPHO1;3. The distinct expression patterns of the PpPHO1 genes in various tissues, such as protonemata, leaves, rhizoids, and auxiliary hairs, suggest that they may play distinct, yet overlapping, functions in Pi transport and/or Pi homeostasis in the various tissues of the moss. Of particular interest in the context of Pi nutrition is the increased abundance of transcripts of PpPHO1;1 and PpPHO1;7 in plants grown under Pi-deficient conditions, but not in plants grown under other nutrient deficiencies. In contrast to Pi deficiency-responsive genes in Arabidopsis, such as IPS1, RNS1, SQD1, and PHT1;1, which are strongly up-regulated within 4 to 24 h after a shift to Pi-deficient conditions (Karthikeyan et al., 2002; Hammond et al., 2003; Misson et al., 2005; Morcuende et al., 2007), the PpPHO1;1 and PpPHO1;7 genes responded more slowly to Pi deficiency, with the first increase in expression being detectable only by 3 to 5 d by northern analysis, and maximal expression attained by 5 to 11 d (Fig. 5A). A similar slow response in gene expression to Pi deficiency was also evident for the PpSQD1 gene, with clear overexpression only after 9 d of growth under Pi-deficient conditions. The rather slow response of PpPHO1;1, PpPHO1;7, and PpSQD1 to Pi deficiency indicates that these genes likely respond to changes in the concentration of internal Pi or Pi-containing metabolites instead of sensing the external Pi concentration. The substantial slower growth of P. patens compared to Arabidopsis could result in a slower decrease in the internal concentration of Pi or Pi-containing metabolites as plants are shifted to Pi-deficient medium. This hypothesis is supported by a comparison of the Pi concentration in P. patens protonemata grown on Pi-deficient medium (Fig. 5B) with similar data obtained for Arabidopsis roots (Lai et al., 2007). Whereas a 45% and 70% decrease in Pi content was observed for Arabidopsis roots 2 and 4 d after a shift to Pi-depleted medium, respectively (Lai et al., 2007), a similar decrease in Pi concentrations for P. patens protonemata was obtained only after 4 and 9 d of Pi starvation (Fig. 4B).
AtPHO1 has been shown to be involved in the transfer of Pi to the root xylem vessel for long-distance transport to the shoot (Poirier et al., 1991; Hamburger et al., 2002). Promoter-GUS analysis of all 11 members of the AtPHO1 gene family has previously revealed that a majority of PHO1 homologs are expressed in the vascular system of various tissues, including roots, leaves, stems, and floral structures (Wang et al., 2004). However, complementation of the pho1 mutant with nine homologs of the Arabidopsis PHO1 gene family showed that only one homolog, namely, AtPHO1;H1, can complement the pho1 mutant, revealing limited functional redundancy of members of the AtPHO1 family in the transfer of Pi to the xylem vessels (Stefanovic et al., 2007). Furthermore, AtPHO1;H4 has been identified as a protein involved in modulating hypocotyl growth under blue light (Kang and Ni, 2006). Together, these results indicate that the biological role of members of the PHO1 family may be quite broad and go beyond the long-distance transport of Pi. In this context, it is interesting to speculate what could be the role and contribution of the members of the PHO1 family of P. patens to Pi homeostasis because mosses lack an elaborate vascular system and their small size and simple body plan reduces the need for an efficient long-distance Pi transport system. Thus, the advantages of P. patens genetics, including the capacity to inactivate several genes by homologous recombination, should make it a valuable plant system to further our understanding of the role of PHO1 proteins in Pi homeostasis and other aspects of plant biology.
Higher plants have been shown to respond to Pi-deficient conditions through numerous changes in gene expression, metabolism, and development (Poirier and Bucher, 2002). In contrast to higher plants, the response of mosses to Pi deficiency has been largely uncharacterized. This work shows that numerous parallels can be found between the response of P. patens and higher plants to Pi deficiency. At the genetic level, in addition to the PpPHO1;1 and PpPHO1;7 genes, transcripts of several P. patens genes whose counterparts in Arabidopsis are known to be up-regulated by Pi deprivation were also increased in abundance following Pi deficiency. These genes included two distinct P. patens homologs to the Arabidopsis +H-Pi cotransporter gene PHT1, one homolog to the AtSQD1 and AtDGD2 genes involved in sulfolipid and galactolipid synthesis, respectively, and one homolog to the AtAPS1 gene encoding an ADP-Glc pyrophosphorylase involved in starch biosynthesis (Fig. 7). A further parallel at the gene expression level is the rapid repression by cytokinin of the increase in expression of genes by Pi deficiency. At the biochemical level, three distinct metabolic adaptations to Pi deficiency found in higher plants, namely, a decrease in the proportion of phospholipids with a corresponding increase in sulfolipids and galactolipids (Essigmann et al., 1998), and an increase in production of acid phosphatases and ribonucleases (Bariola et al., 1994; Coello, 2002), have also been found to occur in P. patens protonemata grown in Pi-deficient medium (Fig. 9).
In higher plants, Pi deficiency induces significant changes in the architecture of the root system, including reduction in root gravitropism, increase of lateral root growth and density, and reduction in primary root growth (Poirier and Bucher, 2002). In addition, Pi deficiency leads to enhanced root hair elongation and density. These changes result in shallower, more profusely branched root systems in Pi-deficient plants, features that are important to increase the acquisition capacity of a nutrient, such as Pi, that is very poorly soluble and immobile in soil. In contrast, deficiency in nitrate, a highly soluble and mobile nutrient that can be leached deep in the soil, results in distinctive changes in root architecture that include increased primary root length and lateral root elongation, whereas the lateral root density, root hair elongation, and density are unchanged (Lynch, 1995). In this perspective, it is interesting to note that the morphological changes of P. patens protonemata to Pi-deficient conditions were strikingly different from Pi-deficient roots of higher plants. Thus, under Pi deficiency, protonemata gave rise to less chloronemata, but more caulonemata that were longer. The frequency of side-branch formation on the caulonemata was also reduced in Pi-deficient conditions (Fig. 8, D and E; Cove, 1992). The combination of these growth modifications resulted in P. patens colonies that had reduced fresh weight but covered a wider surface area (Fig. 8, G and H). In contrast to the root system of higher plants, mosses grow and acquire nutrients on the surface of tree bark, soil, or rocks, areas where the mobility of Pi is relatively high compared to deeper layers of the soil. Thus, growth adaptation of P. patens to Pi deficiency may simply reflect this different Pi mobility, with longer unbranched caulonemata being more adapted to acquire mobile Pi than a more branched structure. These observations suggest that, during evolution, plants first developed strategies to utilize the nutrients present on or near the soil surface, and, as a root system capable of exploring deeper areas of the soil developed, the morphological changes associated with Pi deficiency were adapted to reflect the different dynamics of Pi availability and mobility.
Although rhizoids can be seen as having a similar function to roots of higher plants in anchoring plants to substrates, direct evidence for the implication of rhizoids in nutrient uptake, including Pi, is lacking. Thus, the biological significance of the increase in the length of rhizoids under Pi deficiency and its role in the adaptation of P. patens to Pi deficiency remains to be assessed.
In yeast, adaptation to Pi deficiency implicates coordinated activation of numerous genes under the control of a signal transduction cascade (the PHO regulon) involving numerous proteins, such as kinase (PHO85), kinase inhibitor (PHO81), Pi transporter (PHO84), and transcription factor (PHO4; Ogawa et al., 2000). A similar coordinated response to Pi deficiency also occurs in plants, although the proteins participating in this system are not homologous to the yeast proteins and include the transcription factors PHR1 and WRKY75, the small ubiquitin-like modifier E3 ligase SIZ1, and a E2 conjugase PHO2 (Rubio et al., 2001; Miura et al., 2005; Aung et al., 2006; Bari et al., 2006; Devaiah et al., 2007). Characterization of the response of P. patens to Pi deficiency revealed several common elements between mosses and higher plants to Pi deficiency. Thus, it is likely that some components of the moss Pi deficiency response are homologous to proteins identified in higher plants. In this perspective, it is interesting to note that analysis of the P. patens database revealed a potential homolog to the Arabidopsis PHR1 and SIZ1 (D. Secco and Y. Poirier, unpublished data). However, moss also has morphological responses to Pi deficiency that are distinct from higher plants and that may involve distinct regulatory proteins and mechanisms. P. patens could thus be used as a valuable model system to study the evolution of the response of plants to Pi deficiency.
MATERIALS AND METHODS
Plant Material and Protoplast Transformation
Physcomitrella patens was grown on medium containing NO3 as the main nitrogen source (1.8 mm KH2PO4, pH 6.0, 3.4 mm Ca(NO3)2, 1 mm MgSO4, 45 μm FeSO4, 10 μm H3BO3, 2 μm MnCl2, 0.2 μm CuSO4, 0.2 μm ZnSO4, 0.2 μm CoCl2, 0.1 μm KI, 0.1 μm Na2MoO4) solidified with 0.7% agar in a culture room at 24°C (Ashton et al., 1979). Light was provided by fluorescent tubes under a regime of 16 h light and 8 h darkness. Isolation of protoplasts from fresh materials grown on medium containing NH4 as the main nitrogen source (NO3 medium supplemented with 3 mm NH4 tartrate and 0.5% Glc), polyethylene glycol-mediated transformation, regeneration, and selection of hygromycin- or neomycin-resistant clones were performed as previously described (Schaefer and Zryd, 1997).
For studying gene expression under different nutrient starvation conditions, moss was grown on complete NO3 medium (as the control) and on nutrient-deficient medium for 14 d before collecting tissues for RNA extraction. For Pi-deficient medium, K2SO4 was used in place of KH2PO4 (pH 6.0) and buffered with 1 mm MES (pH 6.0); for NO3−-deficient medium, CaCl2 was used instead of Ca(NO3)2; for SO42−-deficient medium, MgCl2, Fe-EDTA, CuCl2, and ZnCl2 were used instead of MgSO4, FeSO4, CuSO4, and ZnSO4, respectively; for Cl−-deficient medium, CoCl2 was removed and MnSO4 was used to replace MnCl2; for K+-deficient medium, NH4H2PO4 (pH 6.0) and NaI were used in place of KH2PO4 and KI; for Ca2+-deficient medium, KNO3 was used instead of Ca(NO3)2; and for Mg2+-deficient medium, K2SO4 was used in place of MgSO4.
For the time-course expression, plants were first grown on normal NO3 medium for 6 d and then transferred to Pi-deficient medium for various amounts of time before being harvested for RNA extraction.
For studying the effect of cytokinin on gene expression, moss was grown on Pi-deficient NO3 medium for 2 weeks and then transferred in the same medium supplemented with 3 μm 6-benzylaminopurine for 1 to 4 d before harvesting.
cDNA Isolation and Expression Analysis by Northern-Blot and Quantitative PCR
Partial cDNAs for the PpPHO1;4, PpPHO1;5, PpPHO1;6, and PpPHO1;7 genes were amplified from a RT reaction performed on 1 μg of total RNA from protonemata using oligo(dT) and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The oligonucleotides used to amplify the cDNAs were GGACCAATCGCTCATTCTGT and GTAAGCCATCCTGTCCCAGA for PpPHO1;4, ATATTCATGGCAGCGAGTC and GATAATGCGTGCAAATATG for PpPHO1;5, GCTGACCTAACACGCATCAA and ACCCCAATCGACACACAAAT for PpPHO1;6, and ATGGTAGGAAGCCGGAGATT and GACGAAGAAGTCCCCAATCA for PpPHO1;7.
Total RNA was extracted using the TRIzol method (Chomczynski and Sacchi, 1987). Northern blot using gene-specific probes was made using standard procedures (Sambrook and Russell, 2001).
Total RNA from gametophores grown in medium with or without Pi was isolated as described above. Following RT reactions using Moloney murine leukemia virus reverse transcriptase (Promega), real-time PCR was performed using ABsolute QPCR SYBR green mix (ABgene) and the MX3000P real-time PCR system (Stratagene). The gene-specific primers used were 5′-CGGCACCCCTGACACTAACC-3′ and 5′-TCGCTTCGGCTTACGGACTC-3′ for PpPHO1;1, 5′-GAACTTGGGGACGCTCAGGA-3′ and 5′-GTGGCTTTGGCGATGGATTC-3′ for PpPHO1;7, and 5′-TTTCAGCACACTCCCTTCCC-3′ and 5′-AACCATAGTCATCTGCGAAATAAACC-3′ for the P. patens actin gene. Each experiment was repeated twice with independent biological samples and two RT-PCR reactions were performed for each of the samples. Gene expression data were presented relative to average values for the +Pi condition using a relative quantification assay after normalization to the control P. patens actin gene.
Amplification of the Promoter Region of PpPHO1 Homologs
The inverse PCR method (Sambrook and Russell, 2001) was used to amplify the promoter sequences of PpPHO1;2 and PpPHO1;3 homologs. Total genomic DNA of P. patens was digested using two different restriction enzymes (SacI-digested DNA for the amplification of the promoter of PpPHO1;1 and HindIII-digested DNA for the promoters of PpPHO1;2 and PpPHO1;3). Self-circularization reactions were then set up using T4 DNA ligase. Circularized DNA was used as a PCR template for the amplification of the promoter sequence of each gene by two gene-specific primers positioned tail to tail. These oligonucleotides were CAAACTCCCTCTAAATCTC and CGAGGCAACTGGAATTATC for the PpPHO1;2 gene and CAGTGACGTTGTTAAACTC and TCTAACCAATCCTATGTG for PpPHO1;3. The 1,173-bp promoter region of PpPHO1;1 was amplified from genomic DNA using the primers TTAGATTGCGAGCTTTGGTG and GCACACGTAACGACTTGAGG. PCR products were directly cloned in Escherichia coli strain TOP10 using either the pGEM-T easy vector system (Promega) or the GeneJet PCR cloning kit (Fermentas) and sequenced by Microsynth.
Promoter-GUS Construction
Primers with attB1 and attB2 sequences were designed following the guidelines of Gateway technology (Invitrogen) to amplify the cloned promoter region immediately before the start codon of the corresponding gene. PCR and in vitro BP recombination reactions using BP Clonase II enzyme mix and pDONR/Zeo as the donor vector were carried out according to the manufacturer's instructions (Invitrogen). The product of the BP reactions was transformed into competent E. coli cells using heat shock.
The Gateway-compatible plant transformation vector pMDC162 (Curtis and Grossniklaus, 2003), containing the GUS gene placed after the attR2 site, was used as the destination vector. The LR reactions to transfer entry clones to pMDC162 were performed following the manufacturer's instructions (Invitrogen). The products of the LR reactions were used to transform competent E. coli cells using heat shock. The resulting constructs were checked by restriction enzyme digestions and sequencing. For the PpPHO1;1∷GUS construct, the region encompassing the PpPHO1;1 promoter-GUS reporter gene-nopaline synthase terminator was reamplified with oligonucleotides generating XhoI and SalI restriction sites at the 5′ and 3′ ends, respectively, and was further subcloned into the XhoI-SalI sites of the vector pBS108CH-35SNPTb plasmid (Schaefer and Zryd, 1997).
Transgenic P. patens plants were incubated in 100 μL of X-gluc solution (1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronide, 100 mm sodium phosphate, pH 7.0) at 37°C for 6 to 24 h. Fixation was done in 200 μL of 5% (v/v) formalin for 10 min, followed by an incubation for 10 min in 200 μL of 5% (v/v) acetic acid. Pigments in the tissues were removed by serial incubation in 30%, 50%, 70%, and finally 96% ethanol.
Pi Determination and Enzyme Assays
Quantification of Pi in tissues was measured by releasing the cellular content of cells into water by incubation for 45 min at 70°C and quantifying Pi by the molybdate assay (Ames, 1966).
For enzyme assays, P. patens protonemata grown for 14 d on +Pi or −Pi medium was collected and homogenized in extraction buffer containing 20 mm Tris-HCl (pH 8.8), 150 mm NaCl, 1 mm EDTA, 20% glycerol, 1 mm phenylmethylsulfonyl fluoride, 1 μg/mL leupeptin, and 1 mm benzamidine. The homogenate was centrifuged at 6,000g for 15 min to pellet the debris and the supernatant collected. The in-gel phosphatase assay was performed essentially as previously described (Elliot et al., 1986). Briefly, proteins were first migrated on a nondenaturing polyacrylamide gel at 4°C, the gel was then washed three times in 0.1 m acetate buffer (pH 5.0), and phosphatase activity was visualized by 10-min incubation in a solution containing α-naphthyl phosphate (1 mm), Fast Garnet dye (1 mm), in a 100 mm sodium acetate buffer (pH 5.0; Elliot et al., 1986).
Ribonuclease activity was based on the protocol of Yen and Green (1991). Briefly, proteins were separated by SDS-PAGE containing 7 mg/mL of Torulopsis utilis RNA (Sigma). Following electrophoresis, the gel was incubated twice for 10 min in 25% isopropanol (v/v) and 0.01 m Tris-HCl (pH 7), followed by two 10-min washes in 0.01 m Tris-HCl (pH 7), before being incubated for 1 h at 51°C in 0.1 m Tris-HCl. After incubation, gels were washed for 10 min in 0.01 m Tris-HCl and then stained in 0.2% toluidine blue O (Sigma) in 0.01 mm Tris-HCl for 10 min. Gels were destained once for 10 min and twice for 30 min in 0.01 m Tris-HCl.
Lipid Analysis
Lipids from 0.1 g of protonemata grown on +Pi or −Pi medium were extracted with 0.5 mL of chloroform:methanol:formic acid (1:1:0.1 [v/v/v]), followed by addition of 0.25 mL of 1 m KCl, 0.2 m H3PO4. Lipids were separated by thin-layer chromatography as previously described (Dörmann et al., 1995). Individual lipids were identified by exposing a portion of the plate to iodine vapor, individual spots were collected from the plates, and fatty acid methyl esters were prepared with 1 n HCl in methanol at 80°C for 2 h. The methyl esters were quantified by gas chromatography.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ788527 to DQ788529 for PpPHO1;1 to PpPHO1;3, respectively, and EU258928 to EU258931 for PpPHO1;4 to PpPHO1;7, respectively.
Acknowledgments
We wish to express our gratitude to Didier Schaefer for constant help and support, as well as to Younousse Saidi, Andrijia Finka, and Jean-Pierre Zrÿd. We also thank Hatem Rouached for critical reading of the manuscript and Syndie Delessert for technical assistance.
This work was supported by the “Fonds National Suisse de la Recherche Scientifique” (grant no. 3100A0–105874 to Y.P.). Contributions are also acknowledged from the Université de Lausanne and the “Canton de Vaud.”
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yves Poirer (yves.poirier@unil.ch).
References
- Abel S, Ticconi C, Delatorre C (2002) Phosphate sensing in higher plants. Physiol Plant 115 1–8 [DOI] [PubMed] [Google Scholar]
- Ames BN (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8 115–118 [Google Scholar]
- Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144 427–435 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a miR399 target gene. Plant Physiol 141 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari RP, Pant BD, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J 6 673–685 [DOI] [PubMed] [Google Scholar]
- Bucher M, Rausch C, Daram P (2001) Molecular and biochemical mechanisms of phosphorus uptake into plants. J Plant Nutr Soil Sci 164 209–217 [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162 156–159 [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Hurry V, Kleczkowski L (2001) Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 212 598–605 [DOI] [PubMed] [Google Scholar]
- Coello P (2002) Purification and characterization of secreted acid phosphatase in phosphorus-deficient Arabidopsis thaliana. Physiol Plant 116 293–298 [Google Scholar]
- Cove D, Bezanilla M, Harries P, Quatrano R (2006) Mosses as model systems for the study of metabolism and development. Annu Rev Plant Biol 57 497–520 [DOI] [PubMed] [Google Scholar]
- Cove DJ (1992) Regulation of development in the moss, Physcomitrella patens. In V Russo, S Brody, D Cove, S Ottolenghi, eds, Development—The Molecular Genetic Approach. Springer-Verlag, Berlin, pp 179–193
- Cove DJ (2000) The moss, Physcomitrella patens. J Plant Growth Regul 19 275–283 [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot S, Chang CW, Schweingruber ME, Schaller J, Rickli EE (1986) Isolation and characterization of the structural gene for secreted acid phosphatase from S. pombe. J Biol Chem 261 2936–2941 [PubMed] [Google Scholar]
- Essigmann B, Guler S, Narang R, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Decker EL, Reski R (2005) Molecular tools to study Physcomitrella patens. Plant Biol 7 220–227 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J, Bennett M, Bowen H, Broadley M, Eastwood D, May S, Rahn C, Swarup R, Woolaway K, White P (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, Yu J, Song XW, Yi KK (2005) Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ 28 353–364 [Google Scholar]
- Kang X, Ni M (2006) Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A, Varadarajan D, Mukatira U, Paino D'Urzo M, Damsz B, Raghothama K (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Dormann P (2002) DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J Biol Chem 277 1166–1173 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T, Takamiya KI, Ohta H (2006) Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J 47 238–248 [DOI] [PubMed] [Google Scholar]
- Lai F, Thacker J, Li Y, Doerner P (2007) Cell division activity determines the magnitude of phosphate starvation responses in Arabidopsis. Plant J 50 545–556 [DOI] [PubMed] [Google Scholar]
- Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Peña A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation-responsive genes in Arabidopsis. Plant J 24 559–567 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama K, Jain A, Jouhet J, Block M, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud M, Bechtold N, Raghothama K, Nussaume L (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55 727–741 [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan A, Raghothama K, Baek D, Koo Y, Jin J, Bressan R, et al (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari RP, Gibon Y, Zheng K, Datt W, Pant B, Bläsing O, Usadel B, Czechowski T, Udvardi MK, et al (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30 85–112 [DOI] [PubMed] [Google Scholar]
- Muchhal U, Pardo J, Raghothama K (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge S, Rae A, Diatloff E, Smith F (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31 341–353 [DOI] [PubMed] [Google Scholar]
- Ogawa N, DeRisi J, Brown P (2000) New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell 11 4309–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0024, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama K (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50 665–693 [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216 23–37 [DOI] [PubMed] [Google Scholar]
- Reski R (1999) Molecular genetics of Physcomitrella. Planta 208 301–309 [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin A, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaefer D (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53 477–501 [DOI] [PubMed] [Google Scholar]
- Schaefer D, Zryd J (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11 1195–1206 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin H, Dewbre G, Harrison M (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39 629–642 [DOI] [PubMed] [Google Scholar]
- Smith F, Ealing P, Dong B, Delhaize E (1997) The cloning of two Arabidopsis genes belonging to a phosphate transporter family. Plant J 11 83–92 [DOI] [PubMed] [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y (2007) Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J 50 982–994 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y (2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 135 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y, Green PJ (1991) Identification and properties of the major ribonuclease of Arabidopsis thaliana. Plant Physiol 97 1487–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]