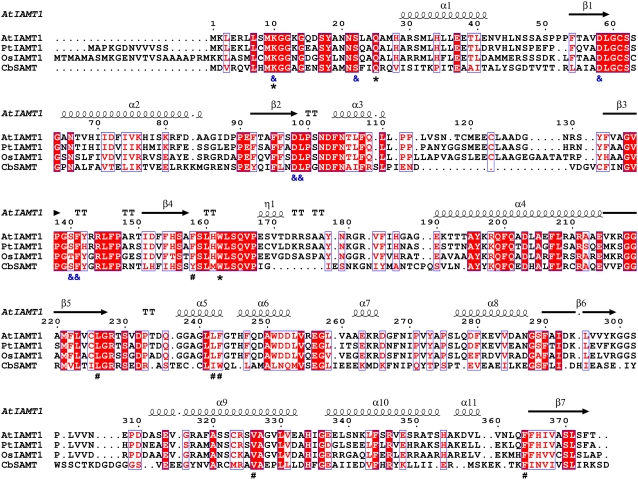
Figure 4.
Structure-based sequence alignment of IAMTs and CbSAMT. Blue frames indicate conserved residues, white characters in red boxes indicate strict identity, and red characters in white boxes indicate similarity. The secondary structure elements indicated above the alignment are those of AtIAMT1, whose structure has been experimentally determined and described here. Residues indicated with “ &” below the alignment are SAM/SAH-binding residues. Residues indicated with “*” are residues that interact with the carboxyl moiety of IAA. Residues indicated with “#” interact with the aromatic moiety of the substrate and are important for substrate selectivity. CbSAMT, C. breweri SAMT (Ross et al., 1999); PtIAMT1, poplar IAMT (Zhao et al., 2007); OsIAMT1, rice IAMT (this study). This figure was prepared with ESPript (Gouet et al., 1999).