Figure 1.
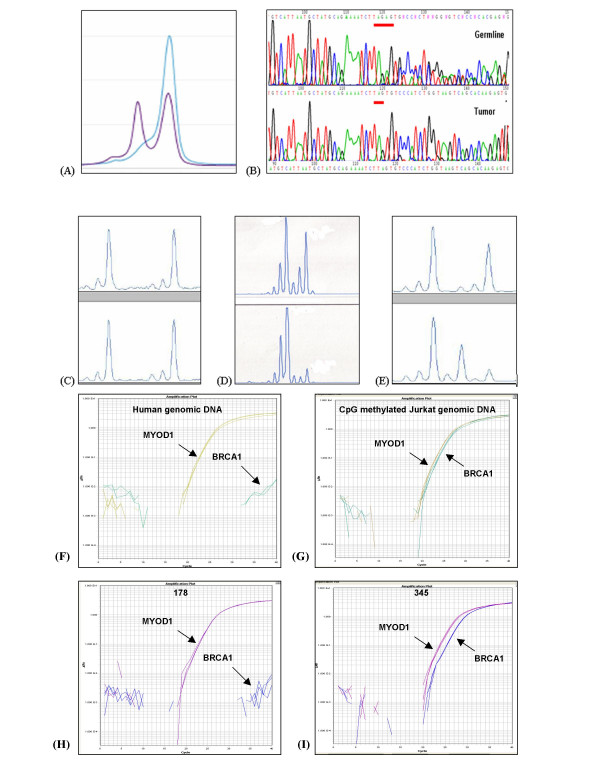
Assessment of BRCA1 loss (A) Mutation screening showing the abnormal denaturing high performance liquid chromatography profile corresponding to the 1351delAT mutation in tumor 223. The single blue line represents the electropherogram from a normal control, while the purple line represents the abnormal profile formed by the mutated exon 11c in tumor 223. (B) Direct DNA sequencing demonstrating the 185delAG mutation in tumor 283. Only the mutant allele is seen in the tumor because LOH is present. (C-E) Loss of heterozygosity (LOH) analysis using BRCA1-associated microsatellite markers visualized on an ABI Prism 3100 Genetic Analyzer, where LOH is defined as >50% decrease in area under the curve when germline DNA (upper tracing) and tumor DNA (lower tracing) are compared. (C) The lack of LOH in tumor 240 demonstrated using microsatellite marker D17S1185, (D) LOH in tumor 283 demonstrated using microsatellite marker D17S855. (E) Microsatellite instability demonstrated in tumor 156 using microsatellite marker D17S1185. (F, G, H, and I) Methylation analysis of BRCA1 gene using fluorescence-based, quantitative, real-time PCR (TaqMan) using SYBR Green 1 as detection method. Two sets of primers, designed specifically for bisulfite converted DNA, were used: a methylated set for the BRCA1 gene and a reference set (MYOD1) to control for input DNA. Specificity of the reactions for methylated DNA were confirmed separately using human genomic DNA (unmethyated; F) and CpG methylated Jurkat genomic DNA (methylated; G), respectively. H and I show representative examples of results from assessment of BRCA1 loss through promoter hypermethyation. Tumor 178 shows only unmethylated BRCA1 promoter, while tumor 345 shows evidence of BRCA1 promoter hypermethylation.