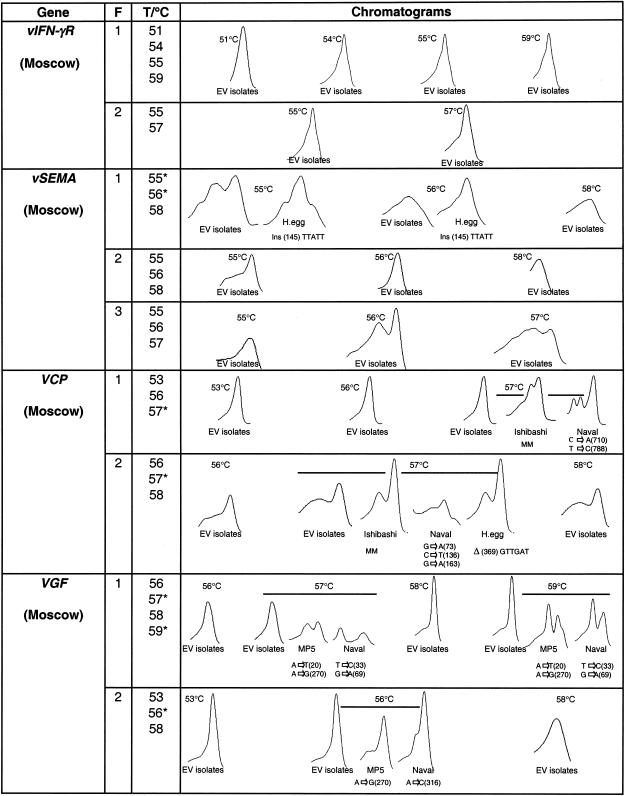
FIG. 2.
DHPLC and sequence analysis of EV genes. The genes were PCR amplified under the following conditions: 30 cycles consisting of 95°C for 1 min, the indicated annealing temperature for 1 min, and 72°C for 1 min, followed by 72°C for 10 min. The PCR fragments were mixed with the reference DNA and subjected to DHPLC analysis. Elution profiles associated with the DHPLC analysis of PCR fragments from the indicated genes are shown. x and y axes depict retention times in minutes after injection and intensity of UV fluorescence at 260 nm, respectively. The fragment number (F) and melting temperature used (T) are indicated, with an asterisk marking those melting temperatures for which mutations were detected. The reference EV used for each gene is shown in parentheses. The results obtained with the reference DNA alone and when it was mixed with DNA from viruses that showed no sequence variation are labeled “EV isolates.” The results from those DNA fragments in which mutations were detected are represented. The base change and the position number of the base affected are given for each chromatogram for samples containing mutations. The presence of multiple mutations (MM), insertions (Ins), and deletions (Δ) is indicated.