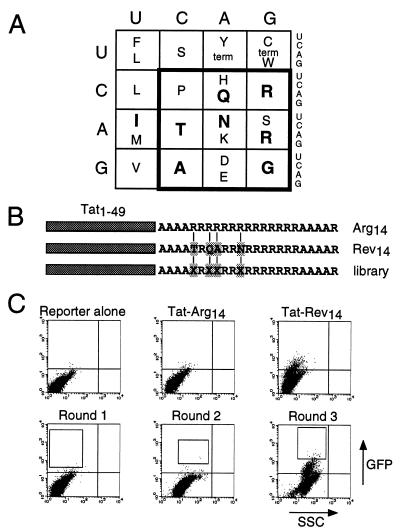
Figure 4.
Screening an arginine-rich combinatorial library for RRE binders. (A) The genetic code viewed from the perspective of arginine-rich peptides. Amino acids known to be important for specific RNA binding by HIV Rev, HIV Tat, BIV Tat, and λ N peptides are indicated in bold (see ref. 24). A restricted genetic code (boldface box) encodes all charged and hydrophilic residues, glycine, alanine, and proline, and contains all six arginine codons. Combinations of these amino acids in an arginine-rich context are expected to encode a variety of helical and nonhelical RNA-binding peptides. (B) Fourteen arginines (Arg14) or a 14-amino acid Rev peptide (Rev14 corresponds to residues 34–47 of Rev, with Trp-45→Arg and Glu-47→Arg substitutions, and specifically binds RRE IIB RNA; ref. 24) were fused to the HIV Tat activation domain (residues 1–49) in the context of surrounding alanines as shown. In the library, four residues corresponding to non-arginine positions in Rev (Xs) were randomized with the amino acids encoded by the boldface box in A. Arg14 served as a negative control and Rev14 as a positive control. (C) FACS analysis of the reporter alone, cells fused to negative and positive control protoplasts used to set sorting windows, and library-containing protoplasts carried through three rounds of sorting. Boxes show the windows used to collect cells in each round. Individual clones from rounds 2 and 3 were tested for activity and positive clones were sequenced.