FIGURE 1. A novel Grb2-deficient cell line.
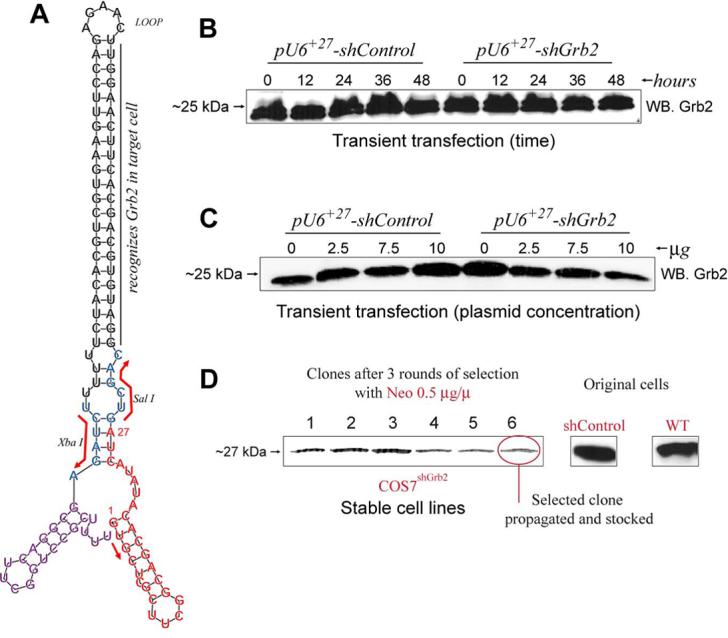
A, Predicted computer-generated stem-loop model using the RNAfold program (Vienna RNA Secondary Structure Prediction program web interface: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) and the shRNA sequence from G+1 to the termination signal UUUU of the RNA polymerase III (position +113) as input. As reference, Sal I and Xba I restriction sites are shown. B, transient transfection of COS7WT cells with 2 mg of pU6+27-shGrb2 or pU6+27-shControl. COS7WT cells were transfected and, at the times indicated, protein samples were obtained in order to monitor endogenous Grb2 protein levels by Western Blot. C, COS7WT cells were transfected with the indicated amounts of pU6+27-shGrb2 or pU6+27-shControl and samples analyzed by Western Blot. D, Generation of stable cell lines. COS7 cells were stably transfected with pU6+27-shGrb2 or pU6+27-shControl as indicated in Methods. Forty clones resistant to neomycin and expressing pU6+27-shGrb2 were screened for Grb2 expression by Western blot. The process was repeated a total of three times. In the end, 3 subclones were stocked and used for further studies. Shown is the expression of Grb2 in comparison with that of original cells.
