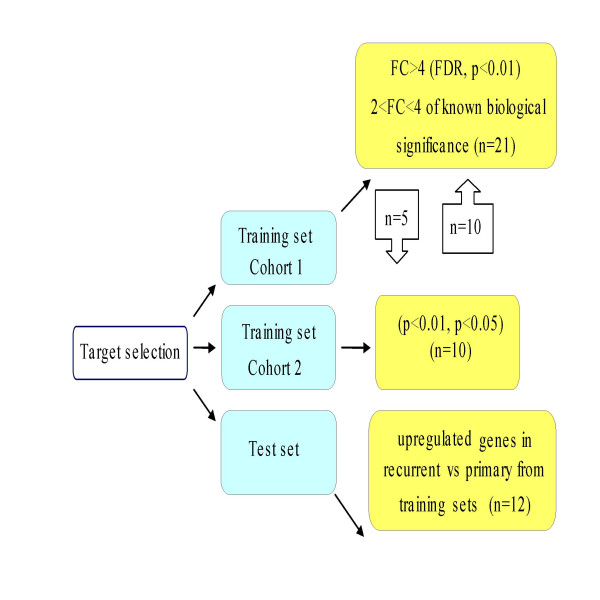
Figure 4.
TaqMan® PCR validation of target genes identified in both training and test sets. Gene selection for TaqMan® validation was based on the most differentially expressed genes from the p and FDR value list with a fold change > 4 but also included genes that had a 2–4 fold change and also some genes involved in the most differentially expressed pathways. Priority was given to selection of genes upregulated in recurrent compared to primary samples, which might provide "recurrence" signatures in ovarian cancer. Upregulated genes validated in both cohorts were alternatively interrogated (external validation) and further advanced for validation in the test set. Independent validation on a test set refers to completely distinct samples of serous histology that were not previously employed in marker development (n = number of gene targets selected for validation).