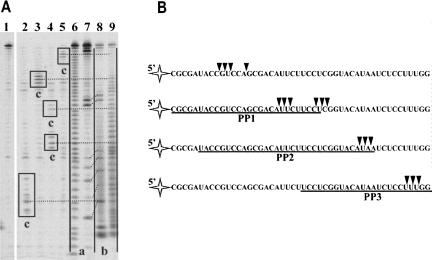
FIGURE 3.
(A) Sequencing of Phi29 DNA polymerase 3′→5′ exoribonuclecleolytic degradation products. The experiments were performed under the conditions described in “Materials and Methods.” The RNA degradation products of 5′-end-labeled free RNA1 or RNA1–PP hybrid substrates, generated by Phi29 DNA polymerase, were analyzed by electrophoresis through sequencing 8% polyacrylamide gel in the presence of RNA ladders. In lane 1 is a control sample of free RNA1. Lanes 2–5 represent the hydrolysis samples of RNA1 in different substrates by Phi29 DNA polymerase (lane 2, free RNA1; lane 3, RNA1-PP2; lane 4, RNA1–PP1; lane 5, RNA1–PP3 hybrids). Lanes 6 and 9 represent RNA1 alkaline hydrolysis ladders before and after the treatment with T4 PNK, respectively. Lanes 7 and 8 represent RNA1 digestion by RNaseT1 (specificity: G) fragments before and after the treatment with T4 PNK, respectively. The products of RNA1 degradation by Phi29 DNA polymerase (lanes 2–5, products c) were aligned with the fragments generated by alkaline hydrolysis and RNaseT1 treatment, and having 3′-phosphate (lanes 6,7, products a) or 3′-OH (lanes 8,9, products b) groups at their 3′ ends. The prevailing RNA1 degradation products are framed. Dotted lines indicate the assignments of bands of RNA1 degradation by Phi29 DNA polymerase to the corresponding bands of alkaline hydrolysis ladder treated with T4 PNK. Oblique dotted lines indicate the assignments of RNA1/RNaseT1 ladder fragments to the same fragments after T4 PNK treatment. (B) Mapping of prevailing RNA degradation products. The underlined sequences indicate areas of target RNA1 hybridization with respective circular DNA probes in RNA–DNA hybrids. Triangles indicate 3′-end points of prevailing RNA oligonucleotides generated by Phi29 DNA polymerase exonuclease activity.