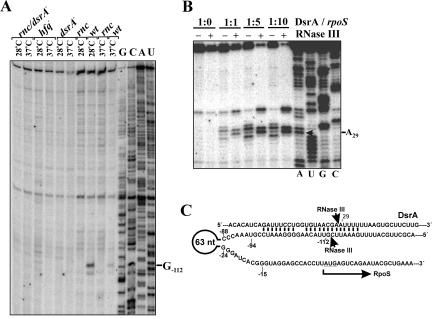
FIGURE 3.
Base-pairing with DsrA induces RNase III cleavage in the rpoS 5′-UTR at an alternative site. (A) Primer extension analysis of total RNA isolated from the wild-type E. coli strain (wt) and isogenic RNase III (rnc), Hfq (hfq), DsrA (dsrA−), and double (rnc/dsrA−) mutants grown at 28°C and 37°C. The product of RNase III cleavage within the rpoS leader (position G−112) accumulates only in the wild-type strain. (B) In vitro synthesized DsrA was incubated alone or with RNase III in the absence (−) or presence (+) of equimolar amount (1:1), fourfold, or ninefold molar excess of RpoSI for 10 min and further analyzed by primer extension. The arrow indicates the nucleotide (A28) at which RNase III cleaves DsrA complexed with RpoSI. (C) Depicted is a part of the rpoS/DsrA complex. Arrows indicate the internucleotide bonds that are cleaved by RNase III. The coordinates of nucleotides in close vicinity to the scissile bonds (G−112 and A29) were determined by primer extension (see Figs. 2B, 3B, respectively).