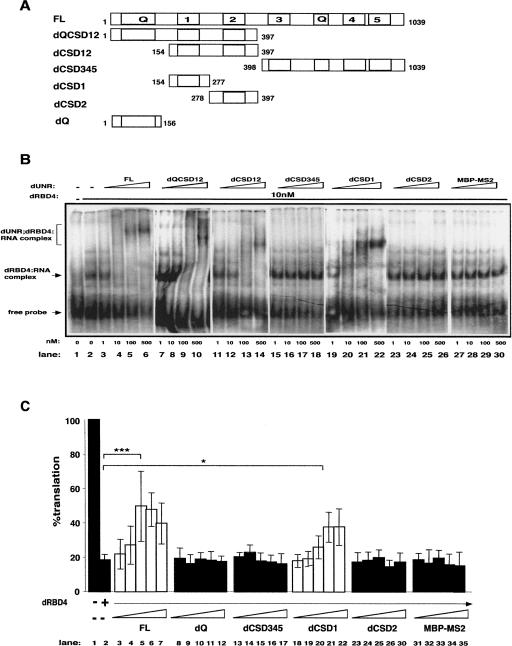
FIGURE 1.
dUNR interacts with SXL and msl-2 mRNA via CSD1. (A) Schematic diagram of dUNR and the deletion derivatives used in this study. dUNR contains five cold-shock domains (1–5) and two Glutamine-rich regions (Q). The positions of the first and last amino acid residues of each construct are indicated. (B) Binding of dUNR and its derivatives to msl-2 mRNA. The 3′UTR EF fragment of msl-2, containing the relevant dUNR and SXL-binding sites, was used in EMSA. Recombinant dUNR fragments were expressed as MBP fusions and were added in increasing amounts (indicated at the bottom) in the absence or presence of 10 nM dRBD4. MBP-MS2 was included as a control. Full-length dUNR (FL) was expressed as a His-tagged fusion. The positions of the different complexes are indicated. (C) Excess recombinant dUNR or dCSD1 derepress translation. BLEF mRNA, containing the minimal msl-2 sequences required for translational repression fused to the Firefly luciferase open reading frame, was used as substrate (see Fig. 3A). The minimal sequences consist of 69 nt in the 5′UTR containing SXL-binding site B, and 46 nt in the 3′UTR containing sites E and F (Gebauer et al. 2003). BLEF mRNA was incubated in typical translation reactions in the absence or presence of 15-fold molar excess of dRBD4, and increasing amounts (0.5, 1, 3, 10, and 30 molar excess over dRBD4) of full-length dUNR or its deletion derivatives. MBP-MS2 was carried as a negative control. Firefly luciferase values were corrected for cotranslated Renilla expression. The activity obtained in the absence of recombinant protein was taken as 100%. The confidence between critical values is indicated (FL, P = 0.00015; dCSD1, P = 0.014).