Abstract
Phenylacetaldehyde dehydrogenase (PAD) and lactaldehyde dehydrogenase (ALD) share some structural and kinetic properties. One difference is that PAD can use NAD+ and NADP+, whereas ALD only uses NAD+. An acidic residue has been involved in the exclusion of NADP+ from the active site in pyridine nucleotide-dependent dehydrogenases. However, other factors may participate in NADP+ exclusion. In the present work, analysis of the sequence of the region involved in coenzyme binding showed that residue F180 of ALD might participate in coenzyme specificity. Interestingly, F180T mutation rendered an enzyme (ALD-F180T) with the ability to use NADP+. This enzyme showed an activity of 0.87 μmol/(min * mg) and Km for NADP+ of 78 μM. Furthermore, ALD-F180T exhibited a 16-fold increase in the Vm/Km ratio with NAD+ as the coenzyme, from 12.8 to 211. This increase in catalytic efficiency was due to a diminution in Km for NAD+ from 47 to 7 μM and a higher Vm from 0.51 to 1.48 μmol/(min * mg). In addition, an increased Kd for NADH from 175 (wild-type) to 460 μM (mutant) indicates a faster product release and possibly a change in the rate-limiting step. For wild-type ALD it is described that the rate-limiting step is shared between deacylation and coenzyme dissociation. In contrast, in the present report the rate-limiting step in ALD-F180T was determined to be exclusively deacylation. In conclusion, residue F180 participates in the exclusion of NADP+ from the coenzyme binding site and disturbs the binding of NAD+.
Keywords: ALDHs, aldehyde dehydrogenases, ALD, lactaldehyde dehydrogenase, catalytic efficiency, burst, coenzyme binding domain
Aldehyde dehydrogenases (ALDHs) oxidize aldehydes to their corresponding acids in reactions coupled to the reduction of NAD(P)+. Most ALDHs are NAD+-dependent enzymes (Perozich et al. 1999; Sophos and Vasiliou 2003), but some use NADP+ (Mann and Weiner 1999; Perozich et al. 2000; Hsu et al. 2001; Cobessi et al. 2000; Ho and Weiner 2005). ALDHs participate in the detoxification of aldehydes, although in a nonspecific manner; therefore, this has been proposed to be their physiological role. However, some ALDHs participate in metabolic pathways or have more specific physiological roles. For instance, ALD and PAD from Escherichia coli K12 participate in the degradation pathways of rare carbon sources (Caballero et al. 1983; Hanlon et al. 1997); betaine aldehyde dehydrogenase is involved in osmolarity regulation (Weretilnyk and Hanson 1990), retinaldehyde dehydrogenase plays a crucial role in cell development and differentiation (Niederreither et al. 2001), and ALDH from Euglena gracilis participates in the energy metabolism of this protist (Rodríguez-Zavala et al. 2006b).
The participation of ALDHs in metabolic pathways is widespread in microorganisms. In E. coli, more than 17 ALDH genes have been identified (Perozich et al. 1999; Sophos and Vasiliou 2003). Three of these enzymes have been characterized recently: aldB, which has been involved in the removal of toxic alcohols and aldehydes in E. coli grown under severe conditions (Ho and Weiner 2005), and ALD and PAD (Rodríguez-Zavala et al. 2006a), which have been identified as part of the degradation pathway of phenylalanine (Hanlon et al. 1997) and of the metabolism of fucose and rhamnose (Caballero et al. 1983; Baldoma and Aguilar 1987; 1988), respectively.
A glutamate at the coenzyme binding domain has been related to the binding and stabilization of NAD+ in NAD+-dependent ALDHs (Perozich et al. 2000). It has been shown that this residue interacts with the 2′-hydroxyl of adenosine ribose. However, a Thr or Lys is found at this position in NADP+-dependent ALDHs (Zhang et al. 1999; Ahvazi et al. 2000); these residues stabilize the 2′-phosphate of NADP+. Paradoxically, a negatively charged residue at this position has also been associated with the exclusion of NADP+ from the active site of NAD+-specific enzymes (Brändén and Tooze 1991). Like most ALDHs, E. coli ALD and PAD possess a Glu residue (E179) in the coenzyme binding site domain, which is involved in the stabilization of the adenine ring. The sequences of PAD and ALD around E179 are almost identical, but PAD can use NAD+ and NADP+ as coenzymes whereas ALD uses only NAD+ (Rodríguez-Zavala et al. 2006a). Therefore, in the present work, site-directed mutagenesis was carried out to examine the role of the E179 region in the determination of the coenzyme preference in E. coli ALD.
Results
Sequence alignment
Sequence alignment of a portion of the coenzyme binding domain around residue E179 (from residue 173 to residue 182) from ALDHs with different coenzyme preference was performed to identify important residue divergences (data not shown). In 10 residues around E179, ALD and PAD sequences are identical except for residue 180, which is Phe in ALD and Thr in PAD (Fig. 1A). As E179 is recognized as an important participant in the binding and stabilization of NAD+ in most ALDHs (Zhang et al. 1999; Perozich et al. 2000), it was judged relevant to determine the orientation and the interactions of F180 in the active site.
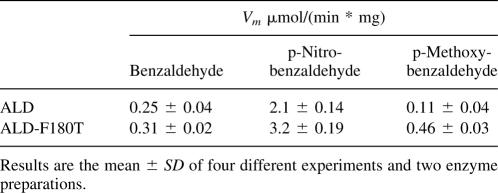
Figure 1.
Representation of the interactions of NADP+ and F180 at the coenzyme binding domain of ALD. Panel A was built by using a homology model obtained for ALD and Panel B was obtained by aligning the three-dimensional model generated for ALD with the published structure of ALD to introduce NADP+ into the coenzyme binding site (Di Costanzo et al. 2007). This figure was generated with PyMOL (http://pymol.sourceforge.net/) and Spdbv (http://www.expasy.org/spdbv/).
Modeling of the tertiary structures of ALD
An ALD three-dimensional model was built by using as template the structure of the E. coli NAD+-dependent medium-chain aldehyde dehydrogenase (1WNB), which showed the highest sequence identity with ALD (38.5%). According to its sequence identity, 1WNB is a member of the betaine aldehyde dehydrogenase family (Gruez et al. 2004). The ALD model was initially generated with NAD+ at the active site (data not shown), as this is the coenzyme found in the 1WNB structure.
Since the interest of this work was to evaluate the inability of ALD to use NADP+, it was imperative to generate a model of the enzyme with this coenzyme at the active site. For that purpose, different structures containing NADP+ were used as templates to model ALD. The template proteins were (1) the three-dimensional structure of ALD generated with 1WNB as the template; (2) two structures of the Streptococcus mutants NADP+-dependent aldehyde dehydrogenase 2euhA (Cobessi et al. 1999) and 1qi1A (Cobessi et al. 2000), which showed 32.3% and 32.6% sequence identity with ALD, respectively; and (3) the structure of the nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase 1ky8A (Pohl et al. 2002), which has 31.7% sequence identity with ALD. The ALD model generated with NADP+ at the active site (Fig. 1A) revealed that residue F180 was at a distance of 3.4 Å from the 2′-phosphate of NADP+, which suffices to perturb its binding. This observation led me to hypothesize that residue F180 may exert a steric effect, which impairs NADP+ binding to the active site, and, thus, this residue affects coenzyme specificity.
The structure of E. coli ALD was recently solved, and two crystals of the enzyme were obtained, one with NADH and another with NADPH in the active site (Di Costanzo et al. 2007). To further explore the F180 interactions, the model here generated was aligned with the ALD crystal structure to introduce NADP+ into the active site (Fig. 1B). This alignment showed that, in the crystal, F180 was at 4 Å from the 2′ phosphate of NADP+ and, thus, it could indeed be exerting a steric effect on this coenzyme at the active site.
Kinetic characterization of ALD-F180T
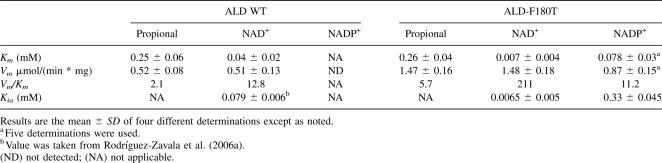
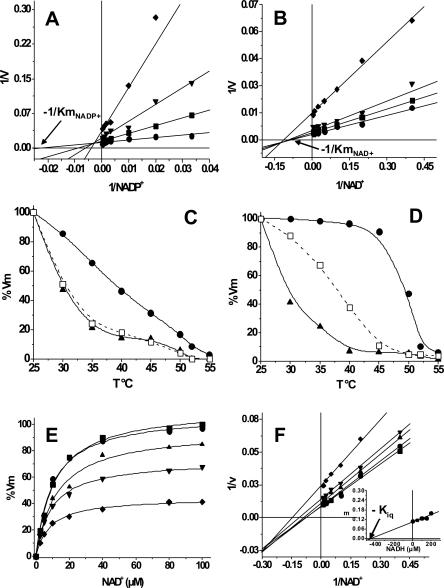
To promote the binding of NADP+ to ALD, residue F180 was mutated to T, as this was the residue found at the same position in PAD, which is able to use both coenzymes (Rodríguez-Zavala et al. 2006a). The mutant protein (ALD-F180T) was purified to homogeneity and the kinetic parameters were determined. Indeed, ALD-F180T showed significant activity with NADP+, which was higher than that attained with NAD+ (Table 1); the Kia (constant related to the Kd for NADP+) value of ALD-F180T for NADP+ revealed high affinity, whereas the Km value for NADP+ was 11 times higher than that for NAD+ (Fig. 2A). Thus, the catalytic efficiency (Vm/Km) of the mutant enzyme with NADP+ was similar to that reached by the wild-type enzyme with NAD+. Surprisingly, Vm of ALD-F180T with NAD+ was threefold higher and Km for NAD+ was sixfold lower than those of ALD wild type (Table 1). Hence, the catalytic efficiency of the mutant with NAD+ was 17-fold higher than that of ALD wild type. The Kia value for NAD+ of ALD-F180T (Fig. 2B; Table 1) was 12-fold lower than that reported for wild-type ALD (79 μM) (Rodríguez-Zavala et al. 2006a), indicating an increase in NAD+ affinity in the mutant.
Table 1.
Kinetic parameters of the mutant ALD-F180T
Figure 2.
Kinetic characterization and thermal stability assays of ALD-F180T. The kinetic parameters were obtained as described in Materials and Methods. Plots shown are representative of five different determinations for NADP+ and four determinations for NAD+ and NADH. (A) Kinetic parameters for NADP+ were obtained by varying propionaldehyde concentration as follows: ●, 2; ■, 0.25; ▼, 0.1; and ◆, 0.05 μM. (B) Kinetic parameters for NAD+ were determined varying propionaldehyde as follows: ●, 2; ■, 0.5; ▼, 0.25; and ◆, 0.05 μM. Km and Kia values obtained from these data are shown in Table 1. (C,D) Protection of ALD and ALD-F180T against thermal denaturalization by the coenzyme. (▲) Enzyme incubated without coenzyme; (□) enzyme incubated with 1 mM NADP+; (●) enzyme incubated with 1 mM NAD+. Activity of ALD incubated with NADP+ was then assayed with 1 mM NAD+. These plots are representative of results obtained with three different enzyme preparations. (E,F) Determination of the Kiq of ALD-F180T. NAD+ kinetics were carried out in the presence of 1 mM propionaldehyde and in the absence (■) or in the presence of the following NADH concentrations: ●, 50; ▲, 100; ▼, 150; and ◆, 200 μM. Plots are representative of four different determinations and two enzyme preparations. The inset in F is the secondary plot of the slopes of the lines against the NADH concentrations. Kiq value represents the mean ± SD of four different determinations. Kiq = 466 ± 60 μM.
Thermal stability studies
Experiments of protection against thermal denaturalization were performed to test whether the lack of activity of wild-type ALD with NADP+ was related to its inability to bind this coenzyme. NADP+ (1 mM) did not protect wild-type ALD against thermal denaturalization (Fig. 2C), suggesting no NADP+ binding, whereas incubation of ALD with 1 mM NAD+ did protect the enzyme. In contrast, NADP+ protected ALD-F180T (Fig. 2C), and this protection was similar to that induced by NAD+ on the ALD wild type (Fig. 2D). Interestingly, the protection against thermal denaturalization by NAD+ was higher in ALD-F180T than in wild-type ALD (Fig. 2D).
Presence of pre-steady-state burst
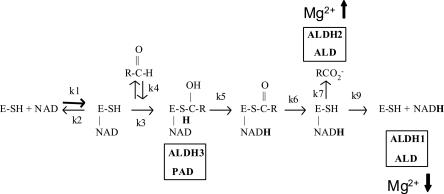
The increase in Vm shown by the mutant enzyme could be related to a change in the rate-limiting step. Pre-steady-state burst analysis was performed with the mutant to determine whether the rate-limiting step was still located after the NADH formation step, as reported for ALD wild type (Rodríguez-Zavala et al. 2006a). ALD-F180T exhibited a pre-steady-state burst (data not shown) indicating that the rate-limiting step remained after the NADH formation (see reaction sequence in Fig. 3). The magnitude of the burst was 2 nmol NADH/nmol enzyme, as determined for ALD wild type (Rodríguez-Zavala et al. 2006a) and for other tetrameric enzymes (Wang and Weiner 1995; Weiner et al. 1976).
Figure 3.
Diagram of the general mechanism of reaction of ALDHs.The rate-limiting step is indicated for some ALDHS: Hydride transfer (k5) is the rate-determining step for ALDH3 and PAD; deacylation (k7) is the rate-limiting step for ALDH3 and partially for ALD; coenzyme dissociation is the rate-determining step for ALDH1 and partially for ALD. Activation by Mg2+ on the deacylation step is indicated by an upward arrow, while Mg2+ inhibiting effect on coenzyme dissociation is indicated by a downward arrow.
Determination of the rate-limiting step
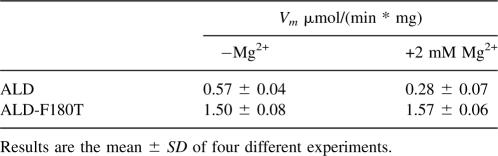
The presence of a pre-steady-state burst discarded the hydride transfer (k5 in Fig. 3) as the rate-limiting step and indicated that this should be located after the formation of NADH, either at the deacylation or the coenzyme dissociation. For ALD wild type, it was previously shown that both deacylation and coenzyme dissociation contribute to limit the reaction rate (Rodríguez-Zavala et al. 2006a). Studies with human enzymes have shown that magnesium affects differentially the kinetic steps of ALDH located after NADH formation. This ion increases the hydrolysis rate of the acyl intermediate (k7 in Fig. 3; Wang and Weiner 1995) and decreases the rate of coenzyme dissociation (k9 in Fig. 3; Takahashi and Weiner 1980; Ho et al. 2005). When the ALD-F180T activity was assayed in the presence of 2 mM Mg2+ (Table 2), no effect on Vm was attained. This Mg2+ concentration inhibits the activity of ALD wild type by 50% (Rodríguez-Zavala et al. 2006a).
Table 2.
Effect of Mg2+ ions on the activity of ALD-F180T
The Kiq constant, a value equivalent to Kd for NADH, was determined to define the modification of the rate-limiting step in the mutant enzyme. The Kiq value determined was 466 ± 60 μM (n = 3) (Fig. 2E,F), a value 3.1-fold higher than the Kiq obtained for ALD wild type (150 ± 27 μM) (Rodríguez-Zavala et al. 2006a). This higher value for Kiq could account for the threefold increase in Vm in the mutant. Furthermore, activity assays with aldehydes with electron-withdrawing and electron-donating substituent groups were made in order to provide an alternative way to assess the rate-limiting step of ALD-F180T. p-Nitrobenzaldehyde was oxidized more rapidly than benzaldehyde or p-methoxybenzaldehyde by ALD-F180T (Table 3), indicating that deacylation was rate limiting.
Table 3.
Activity of ALD-F180T with benzaldehyde and some derivatives
Discussion
Data from the structure of the binary complex of ALD with NADPH indicate that the negatively charged carboxylate group of E179 destabilizes the binding of the 2′-phosphate of NADPH, thus hampering enzyme activity with this coenzyme (Di Costanzo et al. 2007). However, data of the present work (cf. Table 1, Fig. 2) indicate that other factors are involved in the exclusion of NADP+ from the active site. In particular, mutating F180 by T permitted NADP+ binding and utilization, as revealed by the activity exhibited by ALD-F180T with this coenzyme (Table 1) and the improved binding of NAD+ to the enzyme, as indicated by the 6-fold increment in the affinity and the 12-fold diminution in the Kd for this coenzyme as compared to ALD wild-type (Table 1; Fig. 2A,B). Moreover, NADP+ protected the mutant enzyme against thermal denaturalization (Fig. 2D), which was not achieved with wild-type ALD (Fig. 2C). NAD+ protection was higher in ALD-F180T than in wild-type ALD, suggesting that the binding of NAD+ to the mutant enzyme was more efficient than its binding to ALD, which agrees with the lower Km and Kd values of ALD-F180T for NAD+ (Table 1; Fig. 2B). It was surprising that the mutation did not decrease enzyme stability (Fig. 2D). Some reports indicate that single mutations at the coenzyme binding site (Ho and Weiner 2005; Ho et al. 2006) or in regions other than the active site can modify this domain and are accompanied by a detriment in enzyme stability and activity (Rodríguez-Zavala and Weiner 2001, 2002).
The involvement of other factors in the determination of the coenzyme specificity has been demonstrated for other ALDHs. Mutation of T175 of Vh-ALDH to a negatively charged amino acid displaced the preference from NADP+ to NAD+ and increased the catalytic efficiency, while changing this residue to Gln, created a more efficient NAD+-enzyme without loss of NADP+-dependent activity (Zhang et al. 1999). On the other hand, an acidic residue at this position has been associated with the exclusion of NADP+ from the active site of NAD+-specific enzymes (Brändén and Tooze 1991). This glutamate residue is found at position 179 in over two thirds of ALDH sequences, and only some are able to use NADP+ as coenzyme. The ALDH3 family, despite possessing Glu at the active site, can use either NAD+ or NADP+. Mutation of residue E140 of rat ALDH3 (for this enzyme, residue numbering changes, as it does not have 56 amino acids at the N terminus as compared to tetrameric ALDHs; Liu et al. 1997; Perozich et al. 1999; Rodríguez-Zavala and Weiner 2001) to Asn, Asp, Thr, or Gln enhances the preference for NADP+ (Perozich et al. 2000). In addition to E140, which contributes to the tight binding of NAD+, it has been proposed that K137 (highly conserved residue in more than 97% of the ALDH sequences) is essential for binding both coenzymes (Perozich et al. 2000). Nevertheless, in these works, complete abolishment of the use of either coenzyme was unsuccessful, indicating that additional factors contribute to the ability of these enzymes to utilize NADP+.
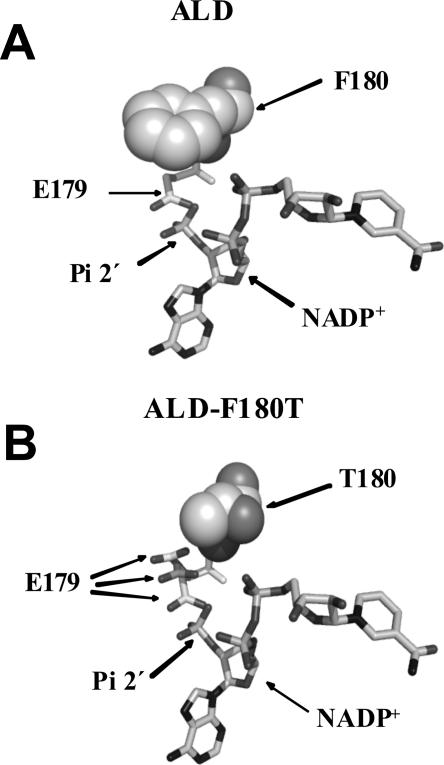
The repulsion effect between the negatively charged groups of NADP+ and E179 is evident from ALD structure data (Di Costanzo et al. 2007). Substituting Phe180 for Thr seems to generate a cavity in the active site, which might allow the side chain of Glu179 to move away from 2′-phosphate of NADP+, thus permitting the adequate coupling of this coenzyme to the active site (Fig. 4B).
Figure 4.
Comparison of the coenzyme binding site of ALD and ALD-F180T. (A,B) F180 and T180 are shown as spheres, while E179 and NADP+ are shown as sticks. E179 is shown in three different positions moving away from E179 and toward T180. Figures were generated using the reported ALD structure (Di Costanzo et al. 2007), with PyMOL (http://pymol.sourceforge.net/) and Spdbv (http://www.expasy.org/spdbv/).
The increase in activity of ALD-F180T with NAD+ with respect to wild-type ALD might be induced by the change in the rate-limiting step, which is shared between deacylation and coenzyme dissociation in ALD wild type. A change in the rate-limiting step was indeed demonstrated by the absence of Mg2+ inhibitory effect on ALD-F180T activity (Table 2). This finding indicates that, for ALD-F180T, NADH dissociation was no longer rate limiting. This observation was further supported by the higher activity obtained with p-nitrobenzaldehyde as compared with that obtained with benzaldehyde (Figs. 3,4; Table 3). The fact that the nitro derivative was oxidized more rapidly is consistent with a nucleophilic attack contributing to the rate-limiting step. Thus, unlike ALD wild type, in ALD-F180T, deacylation is the only rate-limiting step.
Changes in the rate-limiting step by site-directed mutagenesis at the active site of ALDH have been described (Ho et al. 2005, 2006). Mutation of E399 in human ALDH1 changes the rate-limiting step from coenzyme dissociation in ALDH1 wild type (MacGibbon et al. 1977) to hydride transfer in the mutant (Ho et al. 2005), as well as in ALDH3 (Mann and Weiner 1999) and E. coli. PAD (Rodríguez-Zavala et al. 2006a). Furthermore, mutation of T244 to Ser in human ALDH1 rendered an enzyme in which the rate-limiting step changed from coenzyme dissociation to deacylation. The change in rate-limiting step allows changing the inhibitory effect of Mg2+ on ALDH1 wild type to an activating effect on the ALDH1-T244S mutant (Ho et al. 2006).
The increase in NADH Kiq (i.e., Kd = k off/k on) from 150 in the native ALD (Rodríguez-Zavala et al. 2006a) to 466 μM in ALD-F180T (Fig. 3D,E) indicates a faster coenzyme release, and, hence, this step may no longer be rate limiting. Moreover, this increase of about threefold in the Kd of ALD-F180T for NADH could account for the threefold increment in the mutant enzyme Vm.
In general, the NAD+ pool is used by enzymes involved in catabolic pathways, whereas the NADP+ pool is used for biosynthesis. The presence of the Phe residue at the active site of ALD might be a strategy to restrict the use of the NADP+ pool by this enzyme in E. coli. By performing a BLAST analysis with the sequence of E. coli lactaldehyde dehydrogenase, another four ALDHs possessing Phe at position 180 can be found. These enzymes are all found in bacteria that belong to the enterobacteriales (Fig. 5). The sequence identity observed among these enzymes was 83%–85% (data not shown), which indicates that they might share a common ancestor. Therefore, considering the data of the present work (E180 is key for the exclusion of NADP+ from the active site of E. coli ALD, and this residue determines the coenzyme preference in this enzyme), it is predicted that these enzymes are unable to bind and use NADP+ as coenzyme.
Figure 5.
Alignment of the amino acid sequence of the F180 region of ALDHs of some enterobacteriales. The alignment of these sequences showed 83%–85% identity with ALD. Residues E179 and F180 are shown in a rectangle. F180 is in bold lettering.
Materials and Methods
Expression and purification of the recombinant enzymes
The plasmids containing the DNA encoding the ALD protein were kindly provided by Dr. Henry Weiner from the Department of Biochemistry, Purdue University. This sequence was subcloned into a vector to add a His-Tag to the N terminus of the protein to facilitate the purification (Mann and Weiner 1999). Introduction of the mutations was performed by PCR using synthetic oligonucleotides, as described (Ho et al. 1989). E. coli BL21 (DE3)pLysS strain was transformed with the vectors containing the His-Tag constructs. The proteins were overexpressed as reported elsewhere (Rodríguez-Zavala and Weiner 2002). The cells were harvested and washed twice with 100 mL saline solution. Then, the cells were disrupted by sonication at 4°C and the extract was centrifuged at 100,000g for 1 h. The supernatant containing the recombinant protein was applied to a 20-mL Chelating-Sepharose column packed with NiCl2 and equilibrated with a buffer containing 50 mM H2NaPO4 (pH 7.5), 500 mM NaCl, and 20 mM 2-mercaptoethanol at 4°C. The column was washed with 50 mM imidazole in the same buffer, and the protein was eluted applying a 100-mL total volume of a 50–500 mM linear imidazole gradient. Fractions with activity were pooled, concentrated using Amicon filters of 50,000 kDa MWCO, and washed with a buffer containing 100 mM H2NaPO4 (pH 7.5), 100 mM NaCl, and 0.025% 2-mercaptoethanol to eliminate imidazol. The pure enzyme was then concentrated and stored at −20°C in the presence of 50% glycerol until use. Protein stored this way was stable for more than 6 mo. The protein concentration was determined with the bicinchoninic acid protein assay kit (Sigma-Aldrich), using bovine serum albumin as standard. The enzyme purified this way had a purity of more than 95% as judged from SDS-PAGE gels (Laemmli 1970).
Activity assay
For the determination of the activity, 20 μg of protein were added to a buffer containing 100 mM H2NaPO4 (pH 7.5), 100 mM NaCl, 20 mM 2-mercaptoethanol, and 1 mM NAD+ or 1 mM NADP+. The reaction was initiated by the addition of aldehyde. The ALDH activity was measured with the use of a Turner SP-830 spectrophotometer, following the increase in absorbance at 340 nm due to the formation of NADH. The dissociation constants of NADH (Kiq) were determined as inhibition constants using NADH as a competitive inhibitor against NAD+, with propionaldehyde as the substrate. Kms, Kia, and Kiq values were determined from the secondary plots (slope vs. the inverse of coenzyme concentration) of the Lineweaver Burk graphs of the double substrate kinetics shown in Figure 2. In these secondary plots, Kia and Kiq are calculated from the abscissa to the origin. The rate equation for a Bi–Bi ordered reaction is
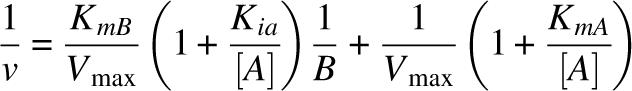 |
where A is NAD+, B is propionaldehyde, KmA is Km for NAD+, KmB is Km for propional, and Kia is Kd for NAD+.
Thermal denaturalization assays
Protein (200 μg) was incubated at the indicated temperature in the absence or in the presence of the coenzyme for 5 min. Then, an aliquot (20 μg) was poured into a cuvette containing the assay buffer at 25°C for activity measurement.
Determination of the pre-steady-state burst
The pre-steady-state burst magnitude of NADH formation was determined by the use of an Aminco Bowman Series 2 spectrofluorometer (SLM-AMINCO Instruments, Inc.) as reported previously (Farrés et al. 1994). The protein (10–20 μM) was incubated in a buffer composed of 100 mM H2NaPO4 (pH 7.4), 100 mM NaCl, and 2 mM NAD+. The reaction was started by the addition of propionaldehyde. The magnitude of the burst of NADH formation was calculated extrapolating the linear portion of the steady-state rate of the reaction to the time of the addition of the substrate. This value was correlated with a calibration curve generated with NADH.
Modeling of the tertiary structures and sequence alignment
Models of the ALD were obtained using the SWISS-MODEL software (available at http://swissmodel.expasy.org/) (Peitsch 1995; Guex and Peitsch 1997; Schwede et al. 2003). Analysis of the structures and generation of the figures were performed with the Protein Explorer program (free software by Erick Martz), the Spdbv software (http://www.expasy.org/spdbv/; Guex and Peitsch 1997), and PyMOL (http://pymol.sourceforge.net/). Sequence alignments were made using the MultiAlign interface by Florence Corpet (Corpet 1988) and the Tcoffe Server (Notredame et al. 2000; Poirot et al. 2003).
Electronic supplemental material
Supplemental Figure 1A shows the alignment of the sequence of the coenzyme binding site of ALDHs with different coenzyme preference and highlights residue F180 in ALD. Supplemental Figure 1B shows the location of F180 with respect to NADP+ in the active site of the ALD model; Supplemental Figure 2 shows the presence of pre-steady-state burst in ALD-F180T mutant. Supplemental Figure 3 shows the cavity formed in the active site of ALD when residue F180 is mutated to T, which facilitates the movement of NADH or NADPH out of the coenzyme binding site.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: José Salud Rodríguez-Zavala, Departamento de Bioquímica, Instituto Nacional de Cardiología, Tlalpan D.F. 14080, Mexico; e-mail: rodjos@cardiologia.org.mx; fax: 52 55 55 73 09 26.
Abbreviations: ALDHs, aldehyde dehydrogenases; ALD, lactaldehyde dehydrogenase; PAD, phenylacetaldehyde dehydrogenase; Vh-ALDH, Vibrio harveyi aldehyde dehydrogenase; ALDH1, class 1 aldehyde dehydrogenase; ALDH3, class 3 aldehyde dehydrogenase.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073277108.
References
- Ahvazi, B., Coulombe, R., Delarge, M., Vedadi, M., Zhang, L., Meighen, E., Vrielink, A. Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: Structural implications for cofactor specificity and affinity. Biochem. J. 2000;349:853–861. [PMC free article] [PubMed] [Google Scholar]
- Baldoma, L., Aguilar, J. Involvement of lactaldehyde dehydrogenase in several metabolic pathways of Escherichia coli K12. J. Biol. Chem. 1987;262:13991–13996. [PubMed] [Google Scholar]
- Baldoma, L., Aguilar, J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: Aerobic–anaerobic regulation of L-lactaldehyde dissimilation. J. Bacteriol. 1988;170:416–421. doi: 10.1128/jb.170.1.416-421.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brändén, C., Tooze, J. Garland; New York: 1991. Introduction to protein structure. [Google Scholar]
- Cabellero, A., Baldoma, L., Ros, J., Boronat, A., Aguilar, J. Identification of lactaldehyde dehydrogenase and glycolaldehyde dehydrodenase as functions of the same protein in Escherichia coli . J. Biol. Chem. 1983;258:7788–7792. [PubMed] [Google Scholar]
- Cobessi, D., Tete-Favier, F., Marchal, S., Azza, S., Branlant, G., Aubry, A. Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutants. J. Mol. Biol. 1999;290:161–173. doi: 10.1006/jmbi.1999.2853. [DOI] [PubMed] [Google Scholar]
- Cobessi, D., Tête-Favier, F., Marchal, S., Branlant, G., Aubry, A. Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans . J. Mol. Biol. 2000;300:141–152. doi: 10.1006/jmbi.2000.3824. [DOI] [PubMed] [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Costanzo, L., Gomez, G.A., Christianson, D.W. Crystal structure of lactaldehyde dehydrogenase from Escherichia coli and interferences regarding substrate and cofactor specificity. J. Mol. Biol. 2007;366:481–493. doi: 10.1016/j.jmb.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrés, J., Wang, X., Takahashi, K., Cunningham, S.J., Wang, T.T., Weiner, H. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. J. Biol. Chem. 1994;269:13854–13860. [PubMed] [Google Scholar]
- Gruez, A., Roig-Zamboni, V., Grisel, S., Salomoni, A., Valencia, C., Campanacci, V., Tegoni, M., Cambillau, C. Crystal structure and kinetics identify Escherichia coli YdcW gene product as a medium-chain aldehyde dehydrogenase. J. Mol. Biol. 2004;343:29–41. doi: 10.1016/j.jmb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Guex, N., Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hanlon, S.P., Hill, T.K., Flavell, M.A., Stringfellow, J.M., Cooper, R.A. 2-Phenylethylamina catabolism by Escherichia coli K-12: Gene organization and expression. Microbiol. 1997;143:513–518. doi: 10.1099/00221287-143-2-513. [DOI] [PubMed] [Google Scholar]
- Ho, K.K., Weiner, H. Isolation and characterization of an aldehyde dehydrogenase encoded by the aldB gene of Escherichia coli . J. Bacteriol. 2005;187:1067–1073. doi: 10.1128/JB.187.3.1067-1073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Ho, K.K., Allali-Hassani, A., Hurley, T.D., Weiner, H. Differential effects of Mg2+ ions on the individual kinetic steps of human cytosolic and mitochondrial aldehyde dehydrogenases. Biochemistry. 2005;44:8022–8029. doi: 10.1021/bi050038u. [DOI] [PubMed] [Google Scholar]
- Ho, K.K., Hurley, T.D., Weiner, H. Selective alteration of the rate-limiting step in cytosolic aldehyde dehydrogenase through random mutagenesis. Biochemistry. 2006;45:9445–9453. doi: 10.1021/bi060718c. [DOI] [PubMed] [Google Scholar]
- Hsu, L.C., Shibuya, A., Yoshida, A. Human stomach aldehyde dehydrogenase cDNA and genomic cloning, primary structure, and expression in Escherichia coli . J. Biol. Chem. 1992;267:3030–3037. [PubMed] [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu, Z.J., Sun, Y.J., Rose, J., Chung, Y.J., Hsiao, C.D., Chang, W.R., Kuo, I., Perozich, J., Lindahl, R., Hempel, J., et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- MacGibbon, A.K., Buckey, P.D., Blackwell, L.F. Evidence for two step binding of reduced nicotinamide-adenine dinucleotide to aldehyde dehydrogenase. Biochem. J. 1977;165:455–462. doi: 10.1042/bj1650455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, C.J., Weiner, H. Differences in the roles of conserved glutamic acid residues in the active site of human class 3 and class 2 aldehyde dehydrogenases. Protein Sci. 1999;8:1922–1929. doi: 10.1110/ps.8.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither, K., Vermot, J., Messaddeq, N., Schuhbaur, B., Chambon, P., Dollé, P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128:1019–1031. doi: 10.1242/dev.128.7.1019. [DOI] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Peitsch, M.C. Protein modeling by E-mail. Biotechnology. 1995;13:658–660. [Google Scholar]
- Perozich, J., Nicholas, H., Lindahl, R., Hempel, J. The big book of aldehyde dehydrogenase sequences. An overview of the extended family. Adv. Exp. Med. Biol. 1999;463:5–52. doi: 10.1007/978-1-4615-4735-8_1. [DOI] [PubMed] [Google Scholar]
- Perozich, J., Kuo, I., Wang, B.C., Boesch, J.S., Lindahl, R., Hempel, J. Shifting the NAD/NADP preference in class 3 aldehyde dehydrogenase. Eur. J. Biochem. 2000;267:6197–6203. doi: 10.1046/j.1432-1327.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- Pohl, E., Brunner, N., Wilmanns, M., Hensel, R. The crystal structure of the allosteric non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeum Thermoproteus tenax . J. Biol. Chem. 2002;277:19938–19945. doi: 10.1074/jbc.M112244200. [DOI] [PubMed] [Google Scholar]
- Poirot, O., O'Toole, E., Notredame, C. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Zavala, J.S., Weiner, H. Role of the C-terminal tail on the quaternary structure of aldehyde dehydrogenases. Chem. Biol. Interact. 2001;130:151–160. doi: 10.1016/s0009-2797(00)00230-1. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Zavala, J.S., Weiner, H. Structural aspects of aldehyde dehydrogenase that influence dimer-tetramer formation. Biochemistry. 2002;41:8229–8237. doi: 10.1021/bi012081x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Zavala, J.S., Allali-Hassani, A., Weiner, H. Characterization of E. coli tetrameric aldehyde dehydrogenases with atypical properties compared to other aldehyde dehydrogenases. Protein Sci. 2006a;15:1387–1396. doi: 10.1110/ps.052039606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Zavala, J.S., Ortíz-Cruz, M.A., Moreno-Sánchez, R. Characterization of an aldehyde dehydrogenase from Euglena gracilis . J. Eukaryot. Microbiol. 2006b;53:36–42. doi: 10.1111/j.1550-7408.2005.00070.x. [DOI] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sophos, N.A., Vasiliou, V. Aldehyde dehydrogenase gene superfamily: The 2002 update. Chem. Biol. Interact. 2003;143–144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., Weiner, H. Magnesium stimulation of catalytic activity of horse liver aldehyde dehydrogenase. Changes in molecular weight and catalytic sites. J. Biol. Chem. 1980;255:8206–8209. [PubMed] [Google Scholar]
- Wang, X.P., Weiner, H. Involvement of glutamate 268 in the active site of human liver mitochondrial (class 2) aldehyde dehydrogenase as probed by site-directed mutagenesis. Biochemistry. 1995;34:237–243. doi: 10.1021/bi00001a028. [DOI] [PubMed] [Google Scholar]
- Weiner, H., Hu, J.H., Sanny, C.G. Rate-limiting steps for the esterase and dehydrogenase reaction catalyzed by horse liver aldehyde dehydrogenase. J. Biol. Chem. 1976;251:3853–3855. [PubMed] [Google Scholar]
- Weretilnyk, E.A., Hanson, A.D. Molecular cloning of a plant betainealdehyde dehydrogenase, an enzyme implicated in adaptation to salinity and drought. Proc. Natl. Acad. Sci. 1990;87:2745–2749. doi: 10.1073/pnas.87.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Ahvazi, B., Szittner, R., Vrielink, A., Meighen, E. Change of nucleotide specificity and enhancement of catalytic efficiency in single point mutant of Vibrio harveyi aldehyde dehydrogenase. Biochemistry. 1999;38:11440–11447. doi: 10.1021/bi991101g. [DOI] [PubMed] [Google Scholar]