Figure 2.—
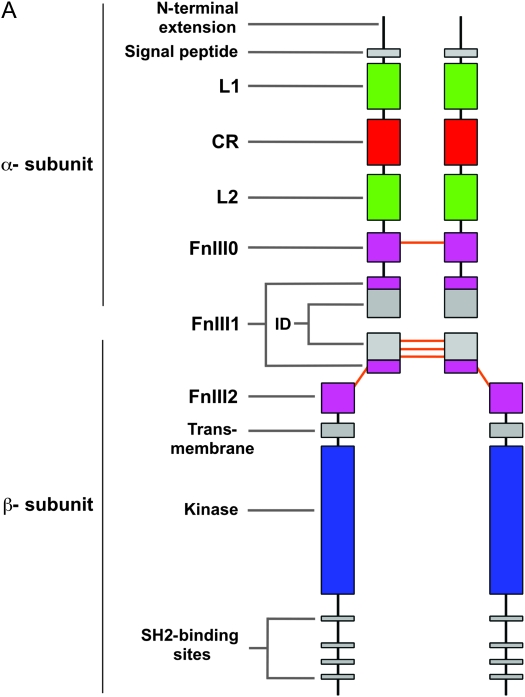
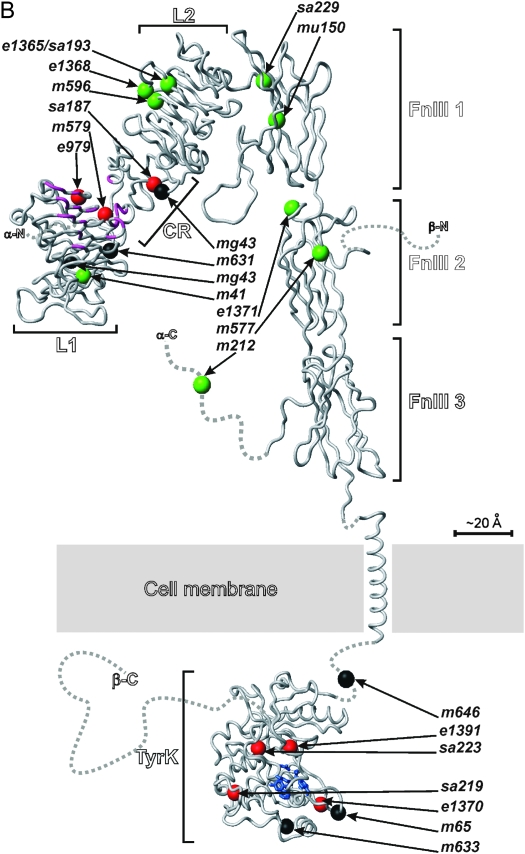
Position and nature of mutant lesions affecting the DAF-2 protein. (A) DAF-2 domain structure. The N-terminal extension and signal peptide are predicted to be cleaved off during processing of the proreceptor. The C-terminal extension is the region C-terminal of the kinase domain. The orange lines represent cysteine residues involved in interchain disulfide bonds. The α- and β-subunits of each heterodimer are linked by a predicted bond between C827, located in the insert domain of FnIII2, and C1126, located within the FnIII3 domain. The holoreceptor is formed by disulfide bonds between equivalent cysteines in adjacent α-subunits at positions C706 (FnIII1), C882, C883, and C886 (insert domain of FnIII2). (B) Backbone representation of homology models of the domains of DAF-2, showing the location of the mutations in the alleles studied. Green, class 1 mutations; red, class 2 mutations; black, nonconditional mutations. The regions of L1 and CR that are thought to interact with human IGF-1 and/or Ins are in purple (note the clustering of class 2 and nonconditional mutations in this region). The active site of the tyrosine kinase domain is in blue. Dashed lines represent regions that are intrinsically unfolded or without known structure. The model was constructed using the atomic coordinates of the extracellular domain of hInsR (PDB code: 2DTG) and the kinase domain of hInsR (PDB code: 1IRK).