Abstract
Previously, we found a novel gene, nuclear receptor interaction protein (NRIP), a transcription cofactor that can enhance an AR-driven PSA promoter activity in a ligand-dependent manner in prostate cancer cells. Here, we investigated NRIP regulation. We cloned a 413-bp fragment from the transcription initiation site of the NRIP gene that had strong promoter activity, was TATA-less and GC-rich, and, based on DNA sequences, contained one androgen response element (ARE) and three Sp1-binding sites (Sp1-1, Sp1-2, Sp1-3). Transient promoter luciferase assays, chromatin immunoprecipitation and small RNA interference analyses mapped ARE and Sp1-2-binding sites involved in NRIP promoter activation, implying that NRIP is a target gene for AR or Sp1. AR associates with the NRIP promoter through ARE and indirectly through Sp1-binding site via AR–Sp1 complex formation. Thus both ARE and Sp1-binding site within the NRIP promoter can respond to androgen induction. More intriguingly, NRIP plays a feed-forward role enhancing AR-driven NRIP promoter activity via NRIP forming a complex with AR to protect AR protein from proteasome degradation. This is the first demonstration that NRIP is a novel AR-target gene and that NRIP expression feeds forward and activates its own expression through AR protein stability.
INTRODUCTION
Classical type I steroid nuclear receptors include androgen receptors (AR), estrogen receptors, progesterone receptors (PR), glucocorticoid receptors (GR) and mineralocorticoid receptors (MR) (1–4). These nuclear receptors function as ligand-inducible transcription factors and typically form ligand-induced homodimers, binding to inverted repeat DNA response elements and then recruit coregulators to promote the expression of target genes (1–4). Coregulators are broadly defined as proteins that play essential roles in the regulation of nuclear receptors. They either enhance transactivation (coactivators) or reduce transactivation (corepressors) of target genes via general transcription factors and chromatin remodeling (5). Over the past decade, several coactivators have been cloned and characterized, ones that associate with steroid receptors and enhance their ability to transactivate target genes (6). Most of these factors enhance assembly of basal transcription factors into a stable pre-initiation complex, resulting in increased transcription initiation rates of RNA polymerase II (7). Previously, we found a novel gene and named it nuclear receptor interaction protein (NRIP) (GenBankTM accession numbers AY766164 and AAX09330). NRIP contains 860 amino acids and seven copies of WD40 domains, and its expression is restricted to the cell nucleus (8). NRIP enhances transcriptional activity of either AR or GR via ligand-dependent interactions (8). We also found that NRIP expression can be induced in prostate cancer cells (LNCaP) treated with androgen. Therefore, whether gene expression of NRIP is induced by hormone is an interesting research question that was investigated in this study.
Regarding prostate cancer, AR plays an important role in male sexual differentiation and prostate cell proliferation (2,9). Prostate-specific antigen (PSA) is a 33 kDa glycoprotein which is elevated in sera from prostate cancer patients (10). We previously characterized that NRIP can enhance an AR-driven PSA promoter activity in LNCaP in a ligand-dependent manner (8). In this study, we extensively investigated the mechanism of regulation of the NRIP gene. Through this investigation, we demonstrate that NRIP represents a novel AR-targeted gene and plays a feed-forward role in enhancing the AR-driven NRIP promoter activity via stabilization of the AR protein. Furthermore, we illustrate that NRIP enhances AR-induced NRIP and PSA gene expression in prostate cancer cells.
MATERIALS AND METHODS
Cell culture and drug treatments
We maintained 293T cells in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml of streptomycin and 100 U/ml of penicillin (Invitrogen). LNCaP cells were grown in RPMI 1640 (Invitrogen) containing 10% FBS, l-glutamine and antibiotics. SL2 cells were cultured in Schneider's Drosophila Medium (Invitrogen) containing 10% FBS and antibiotics. For dihydroxytesterone (DHT) (Sigma-Aldrich) treatment, cells were maintained in the medium containing 5% charcoal–dextran-stripped (CDS) FBS (HyClone) for at least 2 days. For protein stability analysis, cells were treated with 10 µg/ml of cycloheximide (Calbiochem, Darmstadt, Germany) and 10 µM of MG132 (Calbiochem) at the indicated times. Mithramycin A (Sigma-Aldrich) was treated on LNCaP and 293T cells for 24 h at the indicated concentration to inhibit Sp1 binding on DNA (11,12).
Plasmid constructions and site-directed mutagenesis
In reporter constructs, genomic DNA was extracted from HeLa cells using Genomic DNA mini kits (Geneaid, Taoyuan, Taiwan). The region from –2583 to +94 relative to the NRIP transcription start site was amplified by polymerase chain reaction (PCR) with polymerase PfuUltra (Stratagene) and primers NheI-F: 5′-GAGCTAGCAAGGTCAGGGTTGACTT-3′ and HindIII-R: 5′-AAAAGCTTAGGCTCTGCCTGAGC C-3′.
The PCR product was digested with NheI and HindIII and cloned into pGL3-Basic (Promega), and was named NRIP-P2583 (–2583 ∼ +94). NRIP-P413 (–413 ∼ +94) and NRIP-P99 (–99 ∼ +94) were constructed by digestion of XhoI + HindIII and SacI + HindIII from NRIP-P2583, respectively, followed by cloning into pGL3-Basic. A series of 5′ deletion construct fragments of NRIP-P293, NRIP-P258 and NRIP-P234 was amplified by PCR with HindIII-R and forward primers:
293-F, 5′-GAGCTAGCCACACACCAGCCTCA-3′;
258-F, 5′-GAGCTAGCCTCGCGAGAAAGGGT-3′;
234-F, 5′-GAGCTAGCGGGTATCCAGGACGA-3′, respectively.
The amplified fragments were digested by restriction enzymes and constructed on pGL3-Basic. NRIP promoter mutants, NRIP-P413/mARE, NRIP-P413/mGRE and NRIP-P413/mARE/mGRE, were generated using mutation primers,
5′-GACTGTTGCTGATCTTTGGATTTTTTGGTTAG TCTAAGAAGGAGAG-3′ and
5′-GGCTCGGGTGTTGAAACGGGTTTTTTCTCCCC C TCCTCCCCTCCCC-3′.
Mutant constructs NRIP-P413/mSp1-1, NRIP-P413/mSp1-2, NRIP-P413/mSp1-3 were generated by site-directed mutagenesis using mutant primers:
5′-AATGCATTCTTCCAGGGTGAGGAAAGCCGCA GCACACA CCAGCCTC-3′,
5′-TCATCTCGCGAGAAAGGGTTGGAAAGGAGG GTATCCAGGACGAGGA-3′,
5′-CCTCCCCCACGCGGTGGTCTCCAAACCCACC CGGCT CAGGCAGAGCC-3′, respectively. pSG5-HA-Sp1 was a kind gift from Dr Shih-Ming Huang (National Defense Medical Center, Taipei, Taiwan). pPac-hSp1, control vector pPac0 and SL2 cells were kindly provided by Dr Shao-Chun Lu (National Taiwan University, Taipei, Taiwan). pcDNA3.0-AR, pFLAG-AR, pGFP-NRIP, pFLAG-NRIP plasmids were described previously (8). pEBG vector and pEBG DN-Sp1, which is a Sp1-dominant-negative expression plasmid were kind gifts from Dr Thiel (13,14).
RNA interference
pSIN-shNRIP, which encodes short hairpin RNA (shRNA) sequences that knock down the NRIP gene, was generated from EcoRI and KpnI digestion of pSUPER-RNAi-3 as described previously (8) followed by cloning into pSIN-MCS lentiviral vectors. Other RNA interference vectors used in this study were obtained from National RNAi Core Facility (Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan) as follows:
pLKO.1-shAR (target sequence: 5′-CACCAATGTCAA CTCCAGGAT-3′),
pLKO.1-shSp1 (target sequence: 5′-CCAGGTGCAAAC CAACAGATT-3′),
pLKO.1-shGFP (target sequence: 5′-CAACAGCCACAA CGTCTATA T-3′),
pLKO.1-shLuc (target sequence: 5′-CTTCGAAATGTCC GTTCGGTT-3′).
Lentivirus production and infection
Lentiviruses of pSIN and pLKO.1-based vectors were produced by cotransfection with p8.2 and pMD.G into 293FT cells (Invitrogen). Virus titer was usually at 5 × 106 IU/ml. Cells were infected at MOI = 5–10 with 8 μg/ml of polybrene (Sigma-Aldrich).
Transient-transfection and luciferase assays
Transient transfections were performed using calcium phosphate methods for 293T and using Lipofectamine 2000 reagents (Invitrogen) for LNCaP and SL2. For luciferase assays, cells were seeded at a density of 1 × 105 cells/well in 24-well plates. The next day, cells were transfected with reporter constructs and internal controls, pRL-CMV or plasmids, as indicated. Twenty-four hours after transfection, the medium was changed and treated with EtOH or 10 nM DHT. After another 24 h, cells were harvested and luciferase activity was assayed using a Dual-Glo ™ Luciferase Assay System (Promega). The results of the promoter firefly luciferase activities were normalized by internal control Renilla luciferase activities (pRL-CMV or pRL-TK).
RT-PCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen). Five micrograms of RNA was reverse-transcribed by SuperScript ™ III (Invitrogen) and 1 µl of cDNA was amplified by PCR. NRIP, PSA, AR and β-actin primers were as described previously (8).
The primers for the Sp1 gene were as follows:
Sp1 forward: 5′-CTGGTGGGCAGTATGTTGTG-3′,
Sp1 reverse: 5′-AAGCTGGCAGAACTGATGGT-3′, and the amplified fragment was 502 bp.
The primers for the endogenous NRIP gene were as follows:
endoNRIP forward: 5′-GGTGTTGAAACGGGTGT CC-3′
endoNRIP reverse: 5′-ATTGGTGGGCAAATAGC AAC-3′, and the amplified fragment was 688 bp.
The primers for exogenous NRIP-Flag gene expression were as follows:
NRIP-Flag forward: 5′-GGTGCTAACTTTGTAATGA G -3′
NRIP-Flag reverse: 5′-CTTATCGTCGTCATCCTTG T -3′,
and the amplified fragment was 293 bp.
Preparation of cytoplasmic and nuclear extracts
Cells were trypsinized and washed twice with phosphate-buffered saline (PBS). Cell pellets were lyzed in 500 μl of buffer A [10 mM HEPES (pH 7.9), 1.5 mM MgCl2 and 10 mM KCl] with PMSF and DTT, and incubated on ice for 15 min. Insoluble nuclei were separated by centrifugation at 7500g for 5 min at 4°C; the supernatant contained cytoplasmic extracts. The nuclear fraction was washed with buffer A three times and centrifuged at 7500 g for 5 min at 4°C. Pellets (nuclear extracts) were then resuspended in 100 µl of buffer B [20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 420 mM NaCl, 25% (v/v) glycerol and 0.2 mM EDTA] with PMSF, DTT, phospatase inhibitor and protease inhibitors (Sigma-Aldrich), and placed on ice for 20 min.
Co-immunoprecipitation and western blot analysis
Cells were cotransfected with plasmids as indicated. Forty-eight hours after transfection, cell lysates were harvested in NP-40 lysis buffer [150 mM NaCl, 1% NP-40, 50 mM Tris (pH 8.0), 1 mM PMSF and protease inhibitors] and immunoprecipitated with the indicated antibodies as described previously (8). For western blot analysis, proteins were separated on 6% SDS–PAGE, transferred to nitrocellulose membranes, blotted with specific antibodies, and detected using an ECL Western blotting detection system (Amersham Biosciences).
Chromatin immunoprecipitation
Cells were fixed with 1.5% formaldehyde for 15 min at room temperature and then 0.125 M glycine was added to quench cross-linking. Cells were washed 3 times with ice-cold PBS and harvested in 1 ml of cell lysis buffer [5 mM HEPES (pH 8.0), 85 mM KC1, 0.5% NP-40]. Cells were then centrifuged at 5000 r.p.m. at 4°C for 20 min. Nuclear pellets were resuspended in RIPA buffer [10 mM Tris–HC1 (pH 7.8), 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 1 mM PMSF and protease inhibitor] (Sigma-Aldrich). Chromatin was sheared with 10 sets of 10-s pulses on wet ice using a Misonix Sonicator S3000, in which output power was set at level 6. The size of sonicated DNA fragments was between 200 and 500 bp. The soluble chromatin was pre-cleared by Protein G agarose/Salmon Sperm DNA (Upstate) for 1 h at 4°C and then immunoprecipitated overnight at 4°C by an anti-AR antibody (BD PharMingen), anti-Sp1 antibody (Santa Cruz Biotechnology) or anti-NRIP antibody, which was prepared by our lab. Pre-immune normal mouse IgG or rabbit IgG (Santa Cruz Biotechnology) was used as a control. The immunocomplexes were precipitated by Protein G agarose beads for 1 h at 4°C, followed by sequential washing with low salt wash buffer, high salt wash buffer, LiCl wash buffer and TE buffer. For re-ChIP assays, DNA–protein complexes were washed with low salt wash buffer and high salt wash buffer and extracted by adding 10 mM DTT. The supernatants were then diluted 20 times with RIPA buffer for a second-round of immunoprecipitation. After extensive washing, the immunocomplex beads were eluted by elution buffer (1% SDS, 50 mM NaHCO3) for 15 min at room temperature. After reverse cross-linking, proteinase K and RNase A treatments, immunoprecipitated DNA was purified and applied to PCR using primers as follows:
NRIP promoter ARE F: 5′-GACGAGGAGAGGGA GGAGTC-3′,
NRIP promoter ARE R: 5′-GGGACACCCGTTTCAA CAC-3′,
NRIP promoter Sp1-binding site F: 5′-CCGCACAATT CTCTTGCTTC-3′,
NRIP promoter Sp1-binding site R: 5′-GACTCCTCCC TCTCCTCGTC-3′,
PSA promoter ARE as described by Yongfeng Shang et al. (15)
(F: 5′-AGGGATCAGGGAGTCTCACA-3′, R: 5′-GCT AGCACTTGCTG TTCTGC-3′).
The primers for ectopically NRIP promoters to distinguish from endogenous promoter region were as follows:
Luc reporter F: 5′-AGGGTTGGCGGGGAGG GTAT-3′ (containing pGL3-basic vector sequence)
Luc reporter R: 5′-GGAAGACGCCAAAAACATAA AG-3 (containing pGL3-basic vector sequence).
RESULTS
Identification of the human NRIP gene promoter
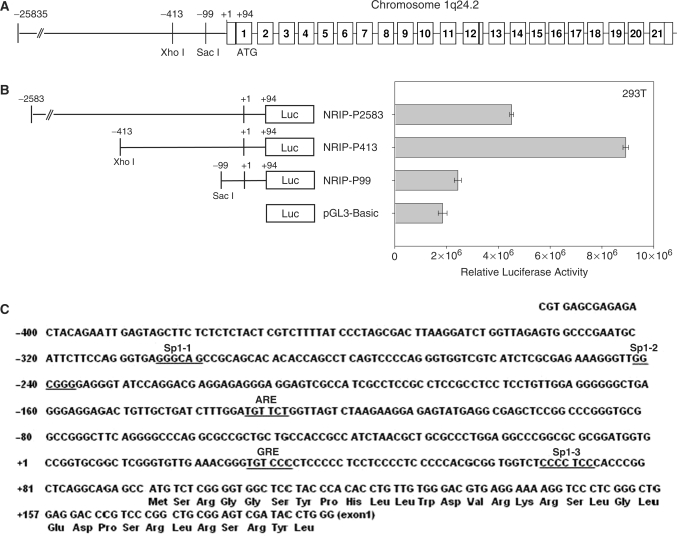
Previously, we isolated a novel gene, NRIP (accession no. AY766164). Expression of this gene can be induced by androgen treatment (8). Human NRIP gene is located at chromosome 1q24 (Figure 1A). The full-length cDNA of NRIP is 3085 bp and contains 93 bp of 5′-untranslated region by 5′-rapid amplification of cDNA ends (RACE)-PCR to determine the transcription start site that is designated as +1 (8). To further investigate the regulation of the NRIP gene, the 5′-flanking region between –2583 and +94 bp upstream from the first exon of the NRIP gene was amplified by PCR from human HeLa cell genomic DNA. The two fragments were generated using restriction enzyme digestion at unique XhoI and SacI sites. These three different region fragments (–2538 ∼+94, –413 ∼+94 and –99 ∼+94) were inserted upstream of the luciferase reporter gene in the pGL3-Basic vector. Transiently transfected into 293T cells, NRIP-P2583 caused about 2-fold increase in luciferase activity relative to pGL3-Basic, but activity was less than that of the NRIP-P413 promoter; and, NRIP-P99 almost lost activity relative to pGL3-Basic activity (Figure 1B). These data suggest that the NRIP core promoter is located at –413 to +94 upstream the initiation site of the NRIP gene.
Figure 1.
Identification of the promoter region in the NRIP gene. (A) Genomic organization of the human NRIP gene on chromosome 1q24.2. The numbers in box refer to exon regions. (B) Identification of the promoter activity in the 5′-flanking region of the NRIP gene. The nucleotides between –2583 and +94 relative to the transcription start site of the NRIP gene were amplified by PCR and cloned into pGL3-Basic and then named NRIP-P2583. Series deletions of ∼ −413 to +94 and ∼ −99 to +94 regions were constructed by XhoI and SacI digestion and cloned into pGL3-Basic and named NRIP-P413 and NRIP-P99, respectively. 293T cells were transiently co-transfected into the indicated reporter promoter with pRL-CMV (as an internal control). The relative luciferase activity is expressed as the measured firefly luciferase activities (promoter activity), which were normalized by renila luciferase activity (pRL-CMV). The results are shown as mean ± SD from three independent experiments. (C) The putative transcription factor binding elements in the NRIP gene. The NRIP promoter sequences (–413 to +94) were analyzed by Transcription Element Search System (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess). Three Sp1 and two hormone response elements, ARE and GRE, were underlined.
We therefore analyzed the NRIP-P413 promoter in the following experiments. DNA sequence analysis utilizing the Transcription Element Search System (TESS) involved cis-elements in the region spanning –413 and +94 bp, which are critical in the control of NRIP gene expression. The NRIP promoter lacked a TATA box and was enriched in GC (Figure 1C). Additionally, the region contained three putative Sp1-binding sites at –305/–300 (Sp1-1), –242/–237 (Sp1-2) and +67/+73 (Sp1-3) and two hormone response elements. An ARE was located at –133/–128 and a glucocorticoid response element (GRE) was present at +28/+33.
NRIP is a novel AR-target gene
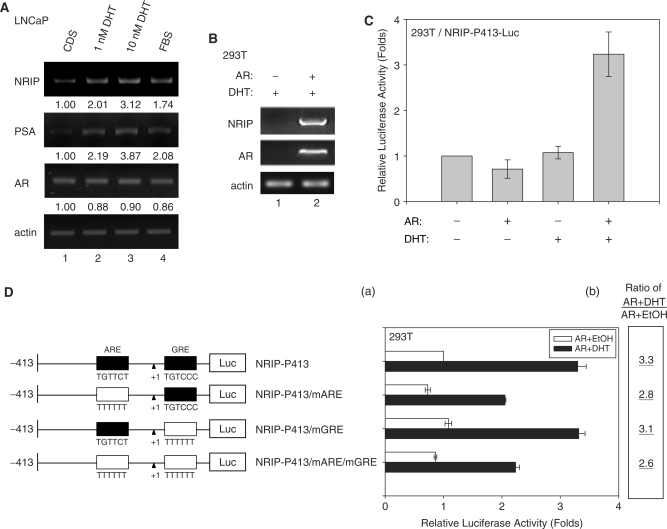
The NRIP promoter contains a potential AR response element based on the sequence analysis; therefore we investigated whether androgen could induce NRIP gene transcription. To examine this hypothesis, prostate cancer cells (LNCaP) were maintained in hormone-depleted medium using CDS serum and then treated with increasing amounts of DHT for 12 h. Total cellular RNAs were subjected to semi-quantitative RT-PCR with NRIP, PSA, AR and β-actin (as an internal control) primers. Since PSA is an AR-targeted gene and a sensitive marker of prostate cancer (4), it is included here as a positive control. As shown in Figure 2A, the expressions of NRIP mRNA in 1 nM DHT and 10 nM DHT are 2.01- and 3.12-fold increases compared to CDS-treated cells, respectively, and the expressions of PSA are 2.19- and 3.87-fold increases. It indicates the expression of NRIP and PSA increased in proportion to the amount of DHT (lanes 2 and 3). NRIP and PSA mRNA were also significantly increased in LNCaP cells cultured in medium with fetal bovine serum (FBS, lane 4) due to a small amount of steroid hormone in the serum compared to CDS. These results indicate that NRIP, like PSA, is an androgen-induced gene. Furthermore, compared with the amount of NRIP mRNA in DHT-treated AR-deficient 293T cells, the NRIP mRNA expression level was elevated when the AR expression plasmid was introduced into cells using DHT treatment (Figure 2B, lane 2). A consistent finding was that the NRIP promoter luciferase activity was increased 3- to 4-fold when 293T cells were transiently cotransfected with NRIP-P413 reporter constructs and AR expression plasmid in the presence of DHT (compared to without AR and DHT; Figure 2C). There was no promoter activity following treatment with DHT alone in 293T cells (Figure 2C). In contrast, the data in Figure 2A showed that DHT alone increased endogenous NRIP gene expression in LNCaP cells. This is due to the fact that the AR gene is rarely expressed in 293T cells but is highly expressed in LNCaP cells (8). These data suggest that the NRIP promoter is activated by androgen-dependent AR activity.
Figure 2.
NRIP is a novel AR-targeted gene. (A) Androgen can induce NRIP and PSA gene expression as measured by RT-PCR in prostate cancer cells (LNCaP). LNCaP cells were grown in RPMI 1640 medium supplemented with 10% FBS or with charcoal/dextran-stripped serum (CDS) and treated with 1 and 10 nM DHT for 24 h. Total RNA was extracted and 5 µg of RNA was amplified by semi-quantitative RT-PCR using NRIP, PSA, AR and β-actin primers. One representative data set from three independent experiments is shown. The expression levels of NRIP, PSA and AR RNA quantified by UVP imaging system were normalized to β-actin, and then set at 1.00 for CDS treatment. (B) AR with androgen can stimulate NRIP gene expression in 293T cells. AR-negative 293T cells were transiently transfected with pcDNA3.0-AR and cultured in CDS medium. After 24 h of 10 nM DHT treatment, total RNA was extracted and RT-PCR was performed using NRIP, AR and β-actin primers. One representative data set from two independent experiments is shown. (C) The promoter activity of NRIP induced by DHT-activated AR. 293T cells were co-transfected with NRIP-P413-Luc and pcDNA3.0-AR or vector (pcDNA3.0) with pRL-CMV and cultured in CDS medium. After 24 h of 10 nM DHT treatment, luciferase activities were measured and normalized. The data are mean ± SD from three independent experiments. The fold change was measured by the luciferase activity of each experimental condition compared to that of the absence of the AR and DHT treatments. (D) AR influences the ARE region of the NRIP promoter. Three site-directed mutants were made by nucleotide substitutions at either ARE or GRE or both sites in NRIP-P413. Wild-type and mutant promoters were transfected with pcDNA3.0-AR and pRL-CMV into 293T cells with or without ligand treatment. The luciferase activities were measured as described above. Panel D(a) depicts the change of relative luciferase activity, which was measured by the luciferase activity of the NRIP-P413 promoter in the absence of AR and DHT treatments. Panel D(b) refers to luciferase activities with DHT relative to that of EtOH for each construct.
To further confirm the importance of an ARE site in NRIP-P-413, we constructed individual site-directed mutant at ARE and GRE or double mutant (16); and named NRIP-P413/mARE, NRIP-P413/mGRE and NRIP-P413/mARE/mGRE, respectively. Figure 2D showed the reduced promoter activity in NRIP-P413/mARE, but the basal level of promoter activity (without ligand treatment) was also lower than that of the wild-type NRIP-P413 promoter. However, the promoter of NRIP-P413/mARE seemed to be affected by DHT treatment [it was increased by 2.8-fold; Figure 2D(b)]. This implies that promoter activity in the mutated ARE within the NRIP promoter can still be affected by androgen. It suggests that there could be additional AR- or androgen-responsive cis-elements. When the promoter NRIP-P413/mGRE in which the GRE site was mutated is stimulated up to 3.1-fold by DHT, implying that the predicted GRE site is not androgen response site. It was further confirmed by the promoter NRIP-P413/mARE/mGRE simultaneously mutated at ARE and GRE sites, and the promoter activity of this double mutant is similar as that of NRIP-P413/mARE. To further support AR involvement in NRIP gene regulation, we used lentiviral vectors (LV), which encode shRNA that target AR. As shown in Figure 3E, lane 2, semi-quantitative RT-PCR results show that shAR can eliminate AR gene expression when we infected LV-shAR into LNCaP cells. The RNA gene expression of NRIP and PSA also significantly decreased in shAR-treated cells compared to the shGFP control (lentivirus encoding shRNA to GFP). In sum, NRIP is a novel AR-target gene.
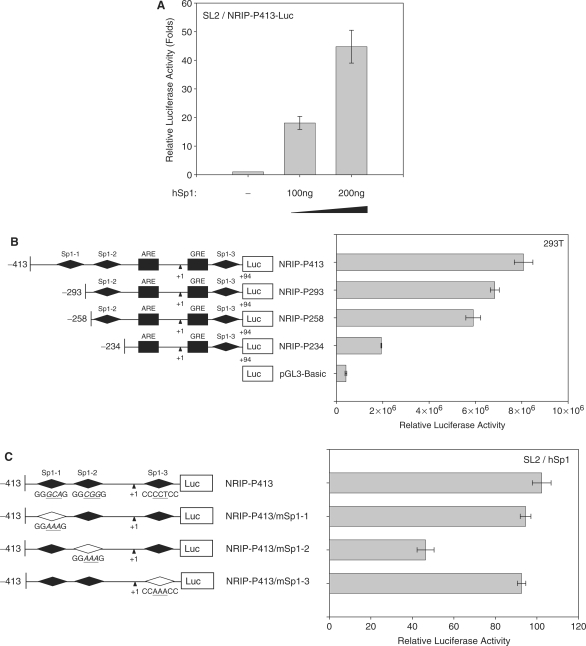
Figure 3.
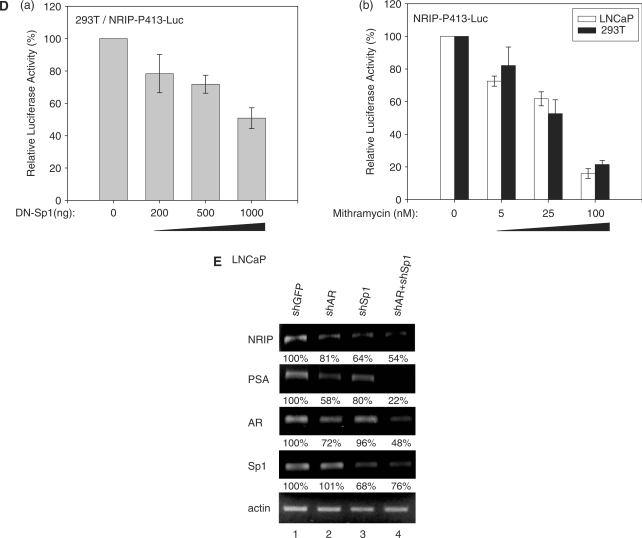
Sp1 regulation on NRIP promoter. (A) Sp1 activation of the NRIP promoter in Drosophila SL2 cells, which lacks endogenous Sp1. Sp1-negative SL2 cells were transfected with NRIP-P413 and increasing amounts of Sp1-expression plasmid (pPac-hSp1). The promoter activity of Sp1 was measured as described above. The fold change was measured by the luciferase activity of each experimental condition compared to that of the absence of the expression of Sp1. (B) NRIP promoter activity measured by a series of Sp1-binding site deletion mutants in 293T cells. A series of 5′-end NRIP-promoter deletion mutants were constructed and named NRIP-P283, NRIP-P256 and NRIP-P234. These lacked Sp1-1, Sp1-1/Sp1-2 and Sp1-1/Sp1-2-binding sites, respectively. Luciferase activities were measured in 293T cells as described in Figure 1B. (C) Sp1 influences the Sp1-2 site of the NRIP promoter. Site-directed mutagenesis at three Sp1-binding sites was generated from the NRIP-P413 promoter and named NRIP-P413/mSp1-1, NRIP-P413/mSp1-2 and NRIP-P413/mSp1-3, respectively. The point mutant sequences were underlined shown in left panel. These three Sp1 site-mutant promoters and pPac-hSp1 were transfected into Sp1-negative SL2 cells. (D) Dominant-negative Sp1 mutant inhibits NRIP promoter activity. Panel a: 293T cells were cotransfected with 0.5 μg of NRIP-P413 and the various doses of the dominant-negative Sp1 expression vector (pEBG DN-Sp1), and the total amount of plasmids was adjusted with empty pEBG vector. The reporter luciferase activity was measured as described above and normalized to the activity of pRL-TK. Panel b: Mithramycin A inhibits NRIP promoter activity. NRIP-P413 was transfected into either LNCaP or 293T cells, 24 h later, cells were incubated with the various concentrations of mithramycine A for another 24 h. Relative NRIP promoter activities (%) were counted as 100% in the cells without treatment of mithramycin A. Data are mean ± SD from three independent experiments. (E) AR and Sp1 cooperative regulation of NRIP transcription. LNCaP cells were infected with lentivirus encoding shRNA to AR and Sp1 individually or in combination; lentivirus-carrying shGFP was a control. Three days post-infection, total RNA was extracted and 5 µg of RNA was amplified by semi-quantitative RT-PCR using NRIP, PSA, AR, Sp1 and β-actin primers. One representative data set from three independent experiments is shown. The expression level of NRIP RNA quantified by UVP imaging was normalized to β-actin.
Sp1 activates NRIP gene expression
There are three Sp1-binding sites in the NRIP-P413 promoter (Sp1-1, Sp1-2 and Sp1-3). To analyze the role of Sp1 transcription factors in the regulation of the human NRIP gene, we introduced various doses of an Sp1-expression plasmid (pPac-hSp1) with an NRIP-P413 promoter into Sp1-deficient Drosophila Schneider SL2 cells (17). Figure 3A showed Sp1 regulating NRIP gene expression in a dose-dependent manner in SL2 cells. To further identify which Sp1-binding site plays the major role in NRIP gene regulation, we generated a 5′ series deletion of NRIP promoter chimeric mutant constructs linked to the luciferase gene. These were named NRIP-P293, NRIP-P258 and NRIP-P234. They lacked Sp1-1, Sp1-1 and Sp1-1/Sp1-2 sites, respectively. Luciferase assays were performed in 293T cells transiently transfected with the indicated promoter. The luciferase activity of NRIP-P234 significantly decreased (Figure 3B), implying that Sp1-2 between –258 and –234 bp is a critical element in the ability of Sp1 to activate NRIP gene expression. To further examine which Sp1-binding sites are functional, we inactivated each Sp1-binding site using site-directed mutagenesis (18). The sites were named NRIP-P413/mSp1-1, NRIP-P413/mSp1-2 and NRIP-P413/mSp1-3, respectively (Figure 3C, left panel). As a result of co-transfection of these mutant constructs with the Sp1 expression plasmid (pPac-hSp1) into SL2 cells, the luciferase activity of NRIP-P-413/mSp1-2 was markedly reduced (Figure 3C). This was consistent with data that the activity of NRIP-P234 was lost in 293T cells (Figure 3B). Taken together, our results demonstrate that Sp1 activates the NRIP promoter through a Sp1-2-binding site.
To further confirm that Sp1 can activate NRIP gene expression, we measured NRIP promoter luciferase activity in the 293T cells cotransfected with the various doses of dominant-negative Sp1 expression plasmid as indicated (13,14). Figure 3D(a) presented that DN-Sp1 inhibited the promoter activity of NRIP in a dose-dependent manner. In addition, mithramycin A is an inhibitor of Sp1 family transcription factors binding to GC-rich promoter regions (11,12), Figure 3D(b) showed that mithramycin A decreased the NRIP promoter activity both in LNCaP and 293T cells in a dose-dependent manner. Moreover, we used LV-shSp1 to knock down endogenous Sp1 expression in LNCaP cells. Semi-quantitative RT-PCR was conducted and RNAs of NRIP, PSA, AR and Sp1 were quantified by a UVP imaging system and normalized by β-actin. The results showed that shSp1 could abolish Sp1 gene expression when infecting LV-shSp1 into LNCaP cells; the expressions of AR, NRIP and PSA mRNAs also decreased in shSp1-treated cells compared to the shGFP control (Figure 3E, lane 3). Since AR and PSA genes are also regulated by Sp1 through Sp1-binding sites in each promoter (2,11), our results are consistent, showing decreases of NRIP and PSA mRNAs by knockdown of Sp1. The amounts of NRIP and PSA RNAs were significantly less after combined shAR and shSp1 treatment compared to single treatment by either shAR or shSp1 (Figure 3E, lane 4). This suggests that either AR or Sp1 can individually regulate NRIP gene expression; and the combination of AR and Sp1 has a synergistic effect in regulating NRIP gene expression.
AR and Sp1 associate on the NRIP promoter and cooperatively regulate NRIP promoter activity
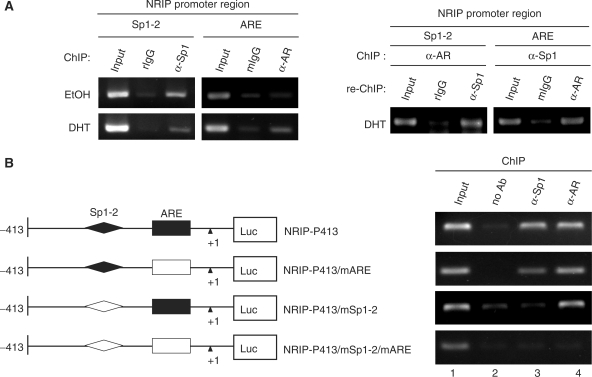
Next, it is important to understand whether AR and Sp1 associate on the NRIP promoter in vivo in the context of chromatin. Chromatin immunoprecipitation (ChIP) was performed using DHT-treated or untreated LNCaP cells. Chromatin extracts were immunoprecipitated with anti-AR or anti-Sp1 antibody and specifically bound DNA was detected by PCR with primer pairs for ARE or Sp1-2-binding sites in the NRIP promoter. As shown in Figure 4A (left panel), the recruitment of Sp1 to Sp1-2 within the NIRP promoter was observed both in the presence and absence of DHT. However, the recruitment of AR to the ARE within the NRIP promoter was identified only in DHT-treated LNCaP cells, indicating Sp1 recruitment to the Sp1-binding site of the NRIP promoter, regardless of androgen. But, AR recruitment to the ARE of the NRIP promoter only occurred after androgen treatment.
Figure 4.
AR and Sp1 associate on the NRIP promoter. (A) The association of AR and Sp1 on the endogenous NRIP promoter in LNCaP cells. Left Panel: ChIP assays were performed in LNCaP cells using primers specific to ARE or Sp1-2-binding sites (as described in Materials and Methods section) to assess AR and Sp1 association with the NRIP promoter with or without DHT. The DNA–protein complexes were then immunoprecipitated by anti-Sp1 and anti-AR and IgG (a negative control) antibodies separately and then subjected to semi-quantitative PCR. rIgG and mIgG represent rat and mouse IgG, respectively. Right Panel: to investigate potential interactions between AR and Sp1 at the NRIP promoter, re-ChIP analysis was performed using the chromatin extracts of the DHT-treated LNCaP cells immunoprecipitated with either anti-AR or anti-Sp1 antibodies. Anti-AR immunoprecipitants and anti-Sp1 immunoprecipitants were re-immunoprecipitated with antibodies to Sp1 or AR, respectively. Then DNAs were extracted and subjected to PCR. (B) The interaction of AR and Sp1 on the ectopic NRIP promoter in 293T cells. 293T cells were transiently transfected with the indicated promoter mutant constructs, pSG5-HA-Sp1 and pcDNA3.0-AR in the presence of DHT. ChIP assays were analyzed by anti-AR and anti-Sp1. (C) Interactions of AR, Sp1 and NRIP. The plasmids pSIN-flag-AR, pSG5-HA-hSp1 and pEGFP-NRIP were co-transfected into 293T cells, which were cultured in FBS medium. After 48 h, cell lysates were collected and immunoprecipitated with anti-GFP, anti-Flag and anti-HA antibodies for NRIP, AR and Sp1, respectively. IP products were subjected to western blotting using antibodies as indicated.
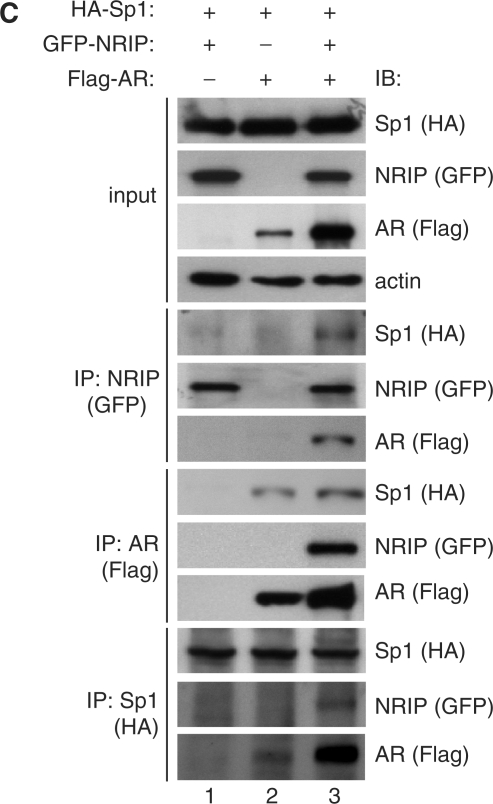
Sp1 is a very important transcription factor, which physically interacts with many cofactors and nuclear receptors to cooperatively activate gene expression (19). The data in Figure 2D show that mutations at the ARE do not completely abolish NRIP promoter activity response to androgen, suggesting that an additional cis-element within the NRIP promoter may be involved in the androgen response. Previous reports have shown that androgen induces p21 promoter activity in which an ARE sequence is absent because an AR can form a complex with an Sp1 protein and AR indirectly binds to Sp1-binding sequences through Sp1 protein (20). Since the NRIP promoter is regulated both by Sp1 and AR, we hypothesized that an interaction between Sp1 and AR is also involved in regulation of the NRIP promoter. Chromatin from DHT-treated LNCaP cells were subjected to re-chromatin immunoprecipitation (re-ChIP) assay. Anti-AR immunoprecipitates were re-immunoprecipitated with anti-Sp1 antibody or control IgG antibody and vice versa. Figure 4A (right panel) shows that Sp1-2 and ARE-containing DNA sequences are both recovered in the anti-AR immunoprecipitates and in the anti-Sp1 immunoprecipitates, respectively, suggesting that AR may bind not only directly to ARE, but also, indirectly to the Sp1-2 element in vivo. Since the distance between ARE and Sp1-2-binding sites is less than 200 bp (Figure 1C), the size of sonicated fragments from chromatin was between 200 and 500 bp under our conditions (data not shown) which hardly distinguishes which transcription factors can bind to each specific DNA-binding site. To verify it, we conducted ChIP assays for AR and Sp1 where the NRIP promoter was ectopically expressed in 293T cells. We constructed several mutant NRIP promoters including NRIP-P413/mARE with a mutated ARE, NRIP-P413/mSp1-2 with mutant at the Sp1-2 site, and NRIP-P413/mSp1-2/mARE containing both the mutated ARE and the mutated Sp1-2. After introducing the indicated promoter with AR- (pcDNA3.0-AR) and Sp1- (pSG5-HA-hSp1) expressing plasmids into 293T cells in the presence of DHT respectively, ChIP analysis was performed and the chromatin extracts from the DHT-treated cells were incubated with anti-AR antibody or anti-Sp1 antibody or without antibodies as a control. As shown in Figure 4B, AR antibody was able to immunoprecipitate chromatin containing ectopic DNAs of NRIP-P413, and NRIP-P413/mARE, and NRIP-P413/mSp1-2 promoters from individually transfected cells (Figure 4B, lane 4). Likewise, Sp1 antibody could immunoprecipitate the DNA of either NRIP-P413 or NRIP-P413/mARE promoter, but not the DNA of the NRIP-P413/mSp1-2 promoter (Figure 4B, lane 3). Neither anti-AR nor anti-Sp1 antibody was able to immunoprecipitate the DNA of NRIP-P413/mSp1-2/mARE promoter with double mutations of ARE and Sp1-2. Taken together, our findings indicate that AR can directly bind to the ARE or indirectly bind to the Sp1-2 element of the androgen-responsive NRIP promoter, but Sp1 only directly binds to the Sp1-2 element and cannot indirectly associate to the ARE through the AR–Sp1 complex.
To further verify AR–Sp1 complex formation, co-immunoprecipitation analysis was performed using transient co-transfection with AR, Sp1 or NRIP expression plasmids (pSIN-Flag-AR, pSG5-HA-hSp1 and pEGFP-NRIP) into 293T cells with DHT treatment. Cell lysates were then collected and immunoprecipitated with anti-GFP, anti-Flag and anti-HA antibodies for NRIP, AR and Sp1, respectively. Figure 4C, lane 2, shows the interactions between AR and Sp1, confirming that AR indirectly binds to the Sp1-2 element of the NRIP promoter via AR–Sp1 complex formation in response to androgen activation. Taken together, like the p21 promoter (20), AR activates the NRIP promoter not only through ARE but also through AR–Sp1 protein complex binding to the Sp1-binding site to induce the androgen response.
NRIP positively autoregulates its own gene expression
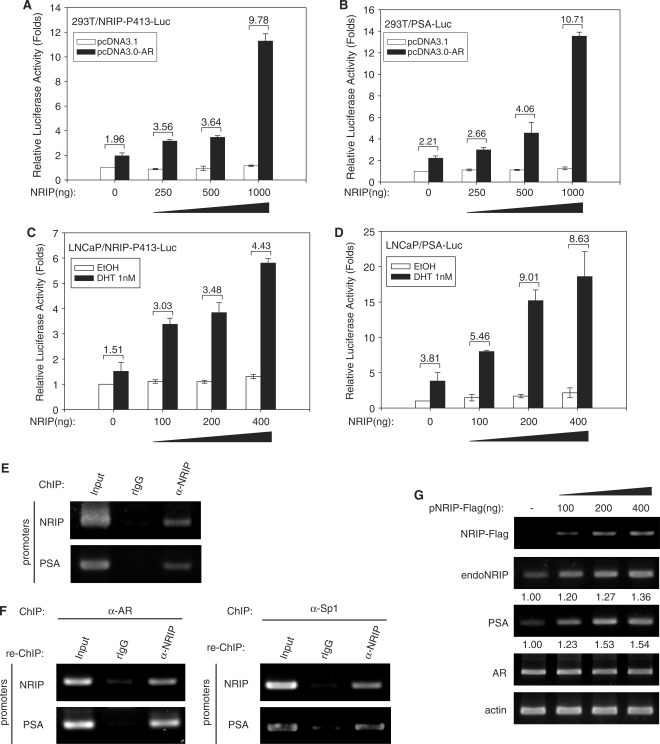
We found that NRIP is a novel AR-target gene. Along with our previous study, this shows that NRIP functions as a transcriptional cofactor to enhance AR-mediated gene expression (8). It raises the interesting question whether NRIP regulates itself via AR. To test the effect of NRIP on its own promoter, we transfected either the NRIP promoter or the PSA promoter (PSA is an AR-target gene and was used as a positive control) with AR- and NRIP-expression plasmids into 293T cells in the presence of DHT. As shown in Figure 5A and B, the enhanced luciferase activities of the NRIP and PSA reporter promoters were correlated with increasing amounts of NRIP gene expression in the presence of DHT, respectively. To further confirm NRIP as an AR transcription cofactor, we introduced the NRIP promoter and the PSA promoter with the indicated amount of NRIP expression plasmid into LNCaP cells which contain endogenous AR. The results confirmed that NRIP activated NRIP and PSA promoter activity in a manner dependent on the NRIP dose (Figure 5C and D). Additionally, we also did ChIP and re-ChIP assays using DHT-treated LNCaP cells to further confirm NRIP-regulating NRIP and PSA promoter activity. Figure 5E shows that NRIP can be complexed with NRIP and PSA promoters. When ChIP was done by either anti-AR or anti-Sp1 antibody; and re-ChIP by anti-NRIP in LNCaP cells in the presence of DHT, the results showed that NRIP could bind to either NRIP or PSA promoter via AR (Figure 5F, left panel) or Sp1 protein (Figure 5F, right panel). It further confirmed ternary complex formation among AR, NRIP and Sp1 as shown in Figure 4C.
Figure 5.
NRIP feed-forward activity enhances its own gene activity in a dose-dependent manner. 293T cells were co-transfected with NRIP-P413-Luc (A) or PSA-Luc (B) with increasing amounts of NRIP expression plasmid, pcDNA3.0-AR, and pRL-CMV (as an internal control). Likewise, LNCaP cells were co-transfected with increasing amounts of NRIP expression plasmid, pRL-CMV, and NRIP-P413-Luc promoter (C) or PSA-Luc promoter (D). Twenty-four hours after transfection, cells were treated with DHT for 12 h and luciferase activities were measured and normalized. The data are mean ± SD from three independent experiments. (E) NRIP can associate with NRIP and PSA promoter by ChIP assay. ChIP assays were performed using DHT-treated LNCaP cells and chromatin extracts were immunoprecipitated by antibodies against NRIP. (F) NRIP associates with either AR or Sp1 to bind to NRIP and PSA promoters, respectively. Chromatin extracts from DHT-treated LNCaP cells were immunoprecipitated by anti-AR or anti-Sp1 antibody and then re-precipitated with antibody against NRIP. DNA was then extracted and amplified by PCR using primers for the ARE region in the NRIP or PSA promoters, separately. (G) LNCaP cells were transiently transfected with increasing amounts of NRIP expression plasmid (NRIP-Flag) and cultured in CDS medium containing 10 nM DHT. After 48 h, cellular RNAs were extracted and subjected to semi-quantitative RT-PCR using primers for NRIP-Flag, endogenous NRIP (endoNRIP), PSA, AR, actin.
In a previous study, we showed an interaction between AR and NRIP proteins (8). To further verify the data of Figure 5C and D, we measured the endogenous mRNA of NRIP and PSA in LNCaP cells by ectopically introduced NRIP-Flag plasmid. Since endogenous NRIP RNA transcripts include a 5′-untranslated region (UTR) covering exon 1 of human NRIP, we amplified this 5′-UTR sequence of endogenous NRIP from total cellular RNA by RT-PCR to distinguish it from the ectopic NRIP-Flag which does not contain the 5′-UTR sequence but has a Flag tag sequence. As shown in Figure 5G, the expression of endogenous NRIP and PSA RNA was correlated with the exogenous amount of NRIP-Flag. Taken together, these findings indicate that NRIP is positively autoregulated by itself.
Complex formation between AR, Sp1 and NRIP were investigated. Co-immunoprecipitation analysis of Figure 4C, lane 1 shows no interaction between Sp1 and NRIP, but complex formation among AR, Sp1 and NRIP did occur (Figure 4C, lane 3). These findings indicate that NRIP through AR indirectly binds to NRIP and PSA promoters and that NRIP plays a role as a mediator in auto-regulation of its own AR-driven expression. Hence, NRIP can cause feed-forward activation of its own promoter activity.
NRIP stabilizes AR protein but has no effect on AR mRNA and AR nuclear translocation
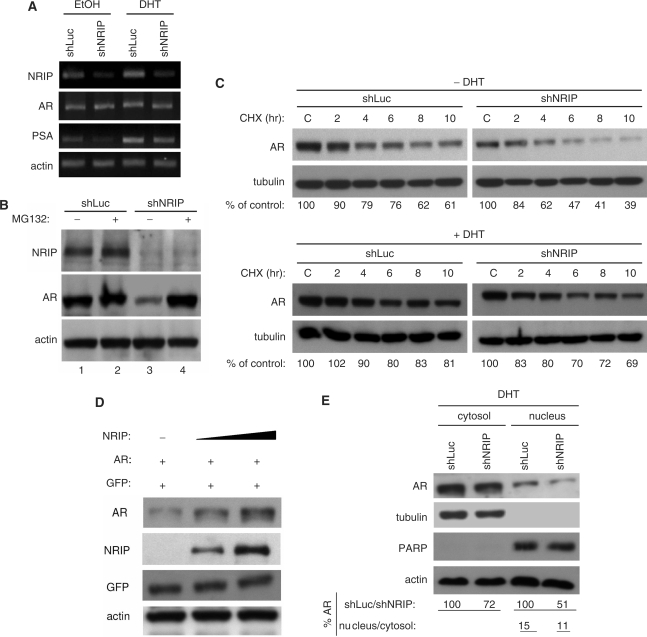
To investigate the positive autoregulatory mechanisms of NRIP, we examined changes in gene expression levels of AR as a result of NRIP effects. We examined the effect of NRIP on AR mRNA, protein levels and subcellular localization. First, we generated recombinant lentiviruses encoding shRNA to NRIP and luciferase (as a control); these were named, respectively, LV-shNRIP and LV-shLuc. LNCaP cells were infected with LV-shNRIP or LV-shLuc. Twenty-four hours post-infection, cells were maintained in CDS medium and treated with EtOH or DHT for 24 h. Total cellular RNA was subjected to semi-quantitative RT-PCR. As shown in Figure 6A, LNCaP cells infected with LV-shNRIP showed decreased NRIP RNA amounts regardless of the presence or absence of DHT, but levels of AR mRNA were maintained at levels seen in cells infected with control virus (LV-shLuc). This implies that knockdown of endogenous NRIP has no effect on the level of AR mRNA.
Figure 6.
NRIP stabilizes AR protein but has no effect on AR mRNA and AR nuclear translocation. (A) The effect of NRIP on AR mRNA. LNCaP cells were infected with LV-shNRIP (or LV-shLuc as control) in the presence or absence of DHT. Three days post-infection, total RNA was extracted and subjected to semi-quantitative RT-PCR using NRIP, AR, PSA or β-actin primers. (B) NRIP effects on proteasome-dependent AR protein degradation. Lentivirus encoding shNRIP infected LNCaP cells in the presence of DHT and with or without proteasome inhibitor (MG132) for 24 h. Cell lysates were analyzed and immunoblotted with anti-NRIP, AR and actin antibodies as indicated. (C) NRIP effects on AR protein stability. LNCaP cells were infected with LV-shNRIP or LV-shLuc. Three days after infection, cycloheximide (CHX) was added for the indicated time. The amounts of AR protein from the lysates of cells in the absence (upper panel) or presence (lower panel) of DHT were analyzed by western blotting using anti-AR and anti-tubulin (as a loading control) antibodies. The% of control indicates the AR amount at each time point relative to the control (without CHX treatment, set at 100). (D) AR stabilization by NRIP in a dose-dependent manner. 293T cells were co-transfected with 1 μg pcDNA3.0-AR, 0, 2 or 4 µg pNRIP-F0lag and 1 µg EGFP-C1 as transfection efficiency controls in a 6-well plate. Total plasmid DNAs were adjusted by pcDNA3.1. Cells were cultured in CDS medium. Forty-eight hours after transfection, cell lysates were subjected to western blotting using antibodies against AR, Flag (for NRIP), GFP and β-actin. (E) The subcellular location of AR by NRIP. LNCaP cells were infected with LV-shLuc and LV-shNRIP for 3 days in the presence of DHT. Cell lysates were separated by cytosol and nuclear fractionation and followed by western blot analysis. The expression level of AR protein was quantified by UVP imaging and normalized by tubulin for the cytosolic fraction and by PARP for the nuclear fraction. For AR stability analysis, proteins from shLuc-treated LNCaP cells were defined as 100%. The percentage of AR nuclear localization was calculated as cytosol/nucleus ratios in shLuc-treated or shNRIP-treated LNCaP cells.
Ubiquitin-dependent proteolysis represents an important mechanism for controlling protein turnover. AR recently was shown to be a target for Mdm2-mediated ubiquitination and destruction by the 26S proteasome (21). To examine whether NRIP can protect AR from proteasome-mediated degradation, LNCaP cells were treated with the proteasome inhibitor MG132 along with infection of LV-shNRIP or LV-shLuc separately. Figure 6B showed AR protein level decreased in compliance with the elimination of NRIP protein in cells treated with shNRIP, but MG132 treatment recovered the AR protein level (Figure 6B, lane 4); indicating that NRIP may modulate AR protein stability via inhibition of proteasome degradation. To further assess whether NRIP regulates the turnover of AR protein, LNCaP cells were infected with LV-shNRIP or LV-shLuc in the presence or absence of DHT. Three days after virus infection, cells were treated with the protein synthesis inhibitor cycloheximide (CHX, 10 μg/ml) at the indicated time. Cell lysates were then extracted to determine AR protein expression levels. We set the AR protein level in each control treatment as 100%, the densitometer measured changes as a percentage of each control treatment. As shown in Figure 6C, the AR protein in shNRIP-treated LNCaP cells were less than those infected with the control shLuc viruses (lane C) both in cells treated with DHT (lower panel) and without DHT (upper panel); indicating that the half-life of the AR protein after shNRIP treatment was significantly shorter than that of the control shLuc regardless of DHT treatment, but AR protein is slightly more stable in cells treated with DHT (lower panel) than without DHT treatment (upper panel). As expected, it previously reported that androgens increase the steady-state expression of AR protein in LNCaP cells (22). This finding of NRIP-stabilizing AR protein is consistent with the result shown in Figure 6B by comparing lane 1 with lane 3. This suggests that AR is subject to proteasome-dependent degradation under normal physiological conditions and that NRIP can stabilize the AR protein. To further confirm a role for NRIP in the stabilization of the AR protein, 293T cells were transiently transfected with the AR expression plasmid and increasing amounts of NRIP expression plasmids. Western blotting analysis showed increasing amounts of AR protein were proportional to NRIP protein levels (Figure 6D), Similarly, as shown in the Input panel of Figure 4C, the amount of AR protein was higher in the presence of GFP-NRIP than the absence (lanes 3 and 2). Taken together, it concludes that NRIP can stabilize the AR protein in a dose-dependent manner.
Because androgen stimulation can induce AR nuclear translocation (1), we were interested in investigating whether knockdown of NRIP would affect the nuclear translocation of AR in the presence of DHT. The fractionation of cytoplasmic and nuclear lysates from LNCaP cells infected with LV-shNRIP or LV-shLuc was done. Figure 6E showed that AR protein amount was decreased in the shNRIP-treated cells both in the fraction of cytosol and nucleus, but no significant difference in the ratio of nuclear to cytosolic proteins between shLuc- and shNRIP-treated LNCaP cells. This indicates that NRIP has no effect on AR nuclear translocation. Taken together, our findings suggest that NRIP stabilizes AR protein but has no effect on AR mRNA and AR nuclear translocation.
DISCUSSION
In this study, we successfully isolated the NRIP promoter, and defined its location and sequence. The core promoter (NRIP-P413) ranged from –413 to 94 bp, from its first exon, which contributes greatly to promoter activity (Figure 1B). Through this investigation, AR and Sp1 were primarily found to influence NRIP gene transcription and the functional AR site at the ARE as well as Sp1 at the Sp1-2 sites within the NRIP core promoter. Our results indicate that NRIP, like PSA, is one of the downstream targets of the AR. Androgen, the natural ligand that activates AR, is essential for the physiological maintenance of the integrity of prostatic epithelial cells (23). Androgen-regulated genes such as PSA (24), p21 (WAF1) (25), KLK-2 (26) have been well characterized, and androgen response elements (AREs) in these genes were identified. AR-target genes are topics that are currently of great interest due to the fact that AR appears to mediate the survival of prostate cancer patients and even of patients with other cancers. Recently, several AR-response genes were found and their functions characterized. For example, fibroblast growth factor 8 (FGF8) is known to induce cell proliferation in human prostate cancer and its presence correlates with tumor stage and pathological grade. FGF8 is induced by AR through ARE in the region of the FGF8 promoter (27). Paternally expressed gene 10 (PEG10) is another gene that is AR-induced through ARE region of the PEG10 gene promoter and upregulated in hepatocellular carcinoma (28). Here we found a novel androgen-regulated gene-NRIP. The correlation of NRIP in prostate cancer development will be interesting for future investigation.
The DNA sequence of the NRIP promoter is TATA-less and GC-rich (Figure 1C). Sp1 is a ubiquitous transcription factor and reportedly binds to GC-rich DNA sequences (GGGCGG) in the proximal promoter regions of a wide variety of genes, especially TATA-less promoters, genes which are involved in a wide range of cellular processes including (a) cell cycle regulation, (b) maintenance of gene activation during embryonic development (by preventing de novo methylation of the CpG islands at house-keeping gene promoters) (19,29) and (c) preservation of chromatin structures at gene loci (30,31). Here we demonstrated that Sp1 stimulates NRIP gene expression at the Sp1-2-binding sequence of the NRIP core promoter. Mutation at the Sp1-2 site caused a reduction in NRIP promoter activity and chromatin immunoprecipitation. Therefore, Sp1 may function as a tethering moiety to recruit the general transcription machinery to a TATA-less NRIP promoter. Previously, Sp1 was reported directly binding to the proximal promoter region of PSA and stimulate the PSA gene expression, implying that Sp1 plays a role in prostate cancer through the regulation of the PSA gene (11). Similarly, here we show that Sp1 also regulates NRIP gene expression (Figure 3) and NRIP was a transcriptional cofactor to enhance AR-mediated PSA gene expression in our previous report (8), indicating that like PSA, NRIP can be a clinical marker of prostate cancer.
Ligand-bound steroid hormone receptors are classically thought to regulate gene expression by binding to the consensus hormone response elements in target genes (32). Intriguingly, when mutation of the ARE did not completely abolish NRIP promoter responsiveness to androgen [Figure 2D(b)], the induction of NRIP gene expression by androgen was mediated not only by an ARE within the NRIP proximal promoter, but also by one Sp1-binding site upon loss of the ARE site (Figure 4B). Similarly, previous studies have shown that androgen activates expression of p21 (WAF1), mouse vas deferens protein (MVDP) and thyroid hormone receptor genes, which contain hormone response elements and Sp1 elements in their promoters (20,33,34). Other than the hormone response element, Sp1 sites in these promoters can respond to hormone action (20). Recently, several studies have also shown that some steroid hormone target genes do not contain the complete hormone response elements within their promoters and can be induced via Sp1 sites by steroid hormone receptor and transcription factor Sp1 interaction. For instance, cathepsin D and heat shock protein 27 (Hsp27) are two estrogen receptor (ER)-target genes and their promoters do not contain consensus estrogen response elements, but have ERE half-sites and GC-rich sequences in their promoters. The ER–Sp1 complex was reported to be necessary for regulation of such promoter activity (35,36). Moreover, cross-talk between nuclear hormone receptors and many other transcription factors has been demonstrated in various studies such as AP1, nuclear factor-kB/Rel, Stat and C/EBP (37,38). Our results further support the notion that hormones induce some of their target genes via the Sp1 site when hormone response elements are absent in these target gene promoters. In sum, we demonstrate that ARE as only half-sites of actual AR-binding sequences and Sp1-2 among three predicted Sp1-binding sites in NRIP core promoter can respond androgen induction. We think that no more additional sequences in NRIP core promoter can be induced by androgen, since the double mutant promoter NRIP-P413/mSp1-2/mARE containing these two mutated sites completely abolishes both AR association (Figure 4B) and luciferase activity (data not shown).
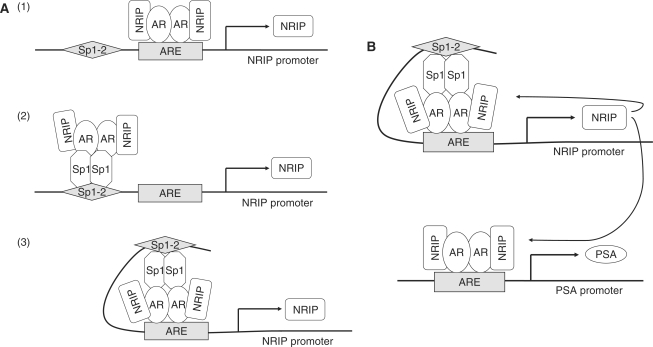
Our study also demonstrates that AR and Sp1 not only bind to their respective consensus sites within the NRIP promoter, but also, the AR–Sp1 complex activates NRIP transcription. This is similar to several reports of promoter regulation by AR and Sp1 such as regulation of p21, TRa and SRY genes (19,20,33). Using protein–protein immunoprecipitation assays, we showed complex formation between AR and NRIP (8) as well as between AR and Sp1 (Figure 4C), but not between Sp1 and NRIP. Therefore, positive regulation of NRIP on AR-target genes at Sp1-binding sequences involves indirect interaction NRIP with Sp1 via AR. On the basis of our results, we propose a functional model of NRIP in which NRIP feeds forward to activate its own promoter activity. This can occur through three possible mechanisms: (i) NRIP protein complexes AR which directly binds to an ARE site; (ii) NRIP forms a complex with the AR-Sp1 protein through the Sp1 protein which binds to the Sp1 site of the NRIP promoter; (iii) The ternary complex bridged through AR and Sp1 tethers to the ARE and Sp1-binding site, resulting in a loop formation on the NRIP promoter (Figure 7A). Formation of this complex in response to androgen may facilitate binding of other coactivators and general transcription factors to form a preinitiation complex for gene transcription. The consequence of this effect would be to enhance expression of the androgen target genes NRIP and PSA (Figure 7B).
Figure 7.
(A) Three proposed models of NRIP gene regulation by AR and Sp1. (1) NRIP protein complexes AR which directly binds to the ARE site; (2) NRIP forms a complex with AR-Sp1 proteins through the AR protein, which indirectly binds to the Sp1 site of the NRIP promoter; (3) A loop forms between ARE and Sp1 sites of the NRIP promoter via the complex formation of AR-Sp1-NRIP. (B) NRIP feed-forward regulation activates its own expression and enhances AR-mediated PSA gene expression.
We showed autoregulatory mechanisms of the NRIP gene, because NRIP can cause AR protein stabilization by avoiding proteasome degradation (Figure 6). Along with our previous study (8), this study shows that NRIP functions as a transcription cofactor to enhance AR-mediated gene expression, which results in the stimulation of NRIP and PSA promoter activity driven by AR (Figure 5). The AR plays a central role in regulating the expression of genes involved in androgen-dependent and androgen-independent tumor formation. Regulation of AR-mediated transcription is achieved by AR protein stabilization and the acetylation status of androgen-responsive genes and/or the AR itself (21). Factors that control AR stability have been confirmed by the finding that the AR is a direct target for Mdm2-mediated ubiquitylation and proteolysis. The E3 ligase activity of Mdm2 and phosphorylation of Mdm2 by PI3K/Akt are essential for Mdm2 to cause AR ubiquitylation and degradation (39). As to how PI3K/AKT affects the AR protein, there are several possibilities. For instance, it was reported that the PI3K/Akt pathway can suppress AR activity in androgen-dependent LNCaP cells with low passage numbers, but enhances AR activity in LNCaP cells with high passage numbers (40). In addition, recent report also demonstrated that PI3K/AKT activation via growth factor (HB-EGF) decreases AR protein levels by regulation of AR mRNA translation rates via mTOR (41). Our study found that NRIP is a cofactor of AR that increases AR transcription activity by stabilizing the AR protein. Further investigations should show what kind of signals is transduced by NRIP effects on the ubiquitin-proteasome pathway of AR, which may aid in the development of drug targets for the treatment of prostate cancer.
NRIP is a transcription cofactor of AR. Most co-activators and co-repressors share a capacity to influence the transcriptional potential of AR by regulating the acetylation–deacetylation status of androgen responsive genes (such as NRIP, PSA) and/or the AR itself, via histone acetyltransferase (HAT) or histone deacetylase (HDAC) activities (chromatin modifying enzymes) that in turn alter the access of AR and general transcription factors to target gene promoters. The co-activators Tip60 (42), p300 and P/CAF (43) enhance the inherent transcriptional activity of the AR by direct receptor acetylation and upregulate transcriptional rates by histone acetylation of AR target genes. Conversely, reversal of HAT activity is important in the deacetylase-dependent abrogation of AR function by histone deacetylase 1 (HDAC1) (15,21,33,42). The further investigation of NRIP complex involving in acetlylation status of AR, may aid in understanding the mechanisms of NRIP role in prostate cancer development.
In sum, NRIP represents a novel AR-targeted gene, prompting an exploration of the correlation between PSA and NRIP, since both are AR-controlled genes. More intriguingly, we demonstrate that NRIP plays a feed-forward role in enhancing the AR-driven NRIP promoter activity via stabilization of the AR protein. Furthermore, we demonstrate that NRIP enhances AR-induced NRIP and PSA gene expression in prostate cancer cells.
ACKNOWLEDGEMENTS
We are indebted to Ms Shu-Chun Lin for technical assistance and thank Dr Zee-Fen Chang, Dr Ming-Shyue Lee, Dr Lih-Hwa Hwang and Dr Shiou-Hwei Yeh for their critical discussion. We thank Dr Gerald Thiel for providing pEBG-Sp1 and pEBG plasmids. This work was supported by National Science Council (NSC 95-2320-B-002-051-MY3) and the National Taiwan University grant (95R0066-BM02-05). RNAi reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taipei, Taiwan and supported by the National Research Program for Genomic Medicine Grants of National Science Council (NSC 94-3112-B-001-003 and NSC 94-3112-B-001-018-Y). Funding to pay the Open Access publication charges for this article was provided by National Science Council of Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black BE, Paschal BM. Intranuclear organization and function of the androgen receptor. Trends Endocrinol. Metab. 2004;15:411–417. doi: 10.1016/j.tem.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Burnstein KL. Regulation of androgen receptor levels: implications for prostate cancer progression and therapy. J. Cell. Biochem. 2005;95:657–669. doi: 10.1002/jcb.20460. [DOI] [PubMed] [Google Scholar]
- 3.Heinlein CA, Chang C. Androgen receptor (AR) coregulators: an overview. Endocr. Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds MA, Kastury K, Groskopf J, Schalken JA, Rittenhouse H. Molecular markers for prostate cancer. Cancer Lett. 2007;249:5–13. doi: 10.1016/j.canlet.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 5.Kumar R, Wang RA, Barnes CJ. Coregulators and chromatin remodeling in transcriptional control. Mol. Carcinog. 2004;41:221–230. doi: 10.1002/mc.20056. [DOI] [PubMed] [Google Scholar]
- 6.Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol. Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 7.Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int. J. Cancer. 2007;120:719–733. doi: 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- 8.Tsai TC, Lee YL, Hsiao WC, Tsao YP, Chen SL. NRIP, a novel nuclear receptor interaction protein, enhances the transcriptional activity of nuclear receptors. J. Biol. Chem. 2005;280:20000–20009. doi: 10.1074/jbc.M412169200. [DOI] [PubMed] [Google Scholar]
- 9.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J. Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 10.Gittes RF. Carcinoma of the prostate. N. Engl. J. Med. 1991;324:236–245. doi: 10.1056/NEJM199101243240406. [DOI] [PubMed] [Google Scholar]
- 11.Shin T, Sumiyoshi H, Matsuo N, Satoh F, Nomura Y, Mimata H, Yoshioka H. Sp1 and Sp3 transcription factors upregulate the proximal promoter of the human prostate-specific antigen gene in prostate cancer cells. Arch. Biochem. Biophys. 2005;435:291–302. doi: 10.1016/j.abb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J. Clin. Invest. 1991;88:1613–1621. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersohn D, Thiel G. Role of zinc-finger proteins Sp1 and zif268/egr-1 in transcriptional regulation of the human synaptobrevin II gene. Eur. J. Biochem. 1996;239:827–834. doi: 10.1111/j.1432-1033.1996.0827u.x. [DOI] [PubMed] [Google Scholar]
- 14.Al-Sarraj A, Day RM, Thiel G. Specificity of transcriptional regulation by the zinc finger transcription factors Sp1, Sp3, and Egr-1. J. Cell. Biochem. 2005;94:153–167. doi: 10.1002/jcb.20305. [DOI] [PubMed] [Google Scholar]
- 15.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 16.Fabre S, Manin M, Pailhoux E, Veyssiere G, Jean C. Identification of a functional androgen response element in the promoter of the gene for the androgen-regulated aldose reductase-like protein specific to the mouse vas deferens. J. Biol. Chem. 1994;269:5857–5864. [PubMed] [Google Scholar]
- 17.Vindevoghel L, Chung KY, Davis A, Kouba D, Kivirikko S, Alder H, Uitto J, Mauviel A. A GT-rich sequence binding the transcription factor Sp1 is crucial for high expression of the human type VII collagen gene (COL7A1) in fibroblasts and keratinocytes. J. Biol. Chem. 1997;272:10196–10204. doi: 10.1074/jbc.272.15.10196. [DOI] [PubMed] [Google Scholar]
- 18.Wang YN, Chang WC. Induction of disease-associated keratin 16 gene expression by epidermal growth factor is regulated through cooperation of transcription factors Sp1 and c-Jun. J. Biol. Chem. 2003;278:45848–45857. doi: 10.1074/jbc.M302630200. [DOI] [PubMed] [Google Scholar]
- 19.Safe S, Kim K. Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:1–36. doi: 10.1016/S0079-6603(04)77001-4. [DOI] [PubMed] [Google Scholar]
- 20.Lu S, Jenster G, Epner DE. Androgen induction of cyclin-dependent kinase inhibitor p21 gene: role of androgen receptor and transcription factor Sp1 complex. Mol. Endocrinol. 2000;14:753–760. doi: 10.1210/mend.14.5.0461. [DOI] [PubMed] [Google Scholar]
- 21.Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeap BB, Krueger RG, Leedman PJ. Differential posttranscriptional regulation of androgen receptor gene expression by androgen in prostate and breast cancer cells. Endocrinology. 1999;140:3282–3291. doi: 10.1210/endo.140.7.6769. [DOI] [PubMed] [Google Scholar]
- 23.Dehm SM, Tindall DJ. Molecular regulation of androgen action in prostate cancer. J. Cell. Biochem. 2006;99:333–344. doi: 10.1002/jcb.20794. [DOI] [PubMed] [Google Scholar]
- 24.Trapman J, Cleutjens KB. Androgen-regulated gene expression in prostate cancer. Semin. Cancer Biol. 1997;8:29–36. doi: 10.1006/scbi.1997.0050. [DOI] [PubMed] [Google Scholar]
- 25.Lu S, Liu M, Epner DE, Tsai SY, Tsai MJ. Androgen regulation of the cyclin-dependent kinase inhibitor p21 gene through an androgen response element in the proximal promoter. Mol. Endocrinol. 1999;13:376–384. doi: 10.1210/mend.13.3.0254. [DOI] [PubMed] [Google Scholar]
- 26.Riegman PH, Vlietstra RJ, van der Korput JA, Brinkmann AO, Trapman J. The promoter of the prostate-specific antigen gene contains a functional androgen responsive element. Mol. Endocrinol. 1991;5:1921–1930. doi: 10.1210/mend-5-12-1921. [DOI] [PubMed] [Google Scholar]
- 27.Gnanapragasam VJ, Robson CN, Neal DE, Leung HY. Regulation of FGF8 expression by the androgen receptor in human prostate cancer. Oncogene. 2002;21:5069–5080. doi: 10.1038/sj.onc.1205663. [DOI] [PubMed] [Google Scholar]
- 28.Jie X, Lang C, Jian Q, Chaoqun L, Dehua Y, Yi S, Yanping J, Luokun X, Qiuping Z, et al. Androgen activates PEG10 to promote carcinogenesis in hepatic cancer cells. Oncogene. 2007;26:5741–5751. doi: 10.1038/sj.onc.1210362. [DOI] [PubMed] [Google Scholar]
- 29.Zhou T, Chiang CM. Sp1 and AP2 regulate but do not constitute TATA-less human TAF(II)55 core promoter activity. Nucleic Acids Res. 2002;30:4145–4157. doi: 10.1093/nar/gkf537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 31.Hung JJ, Wang YT, Chang WC. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol. Cell. Biol. 2006;26:1770–1785. doi: 10.1128/MCB.26.5.1770-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geserick C, Meyer HA, Haendler B. The role of DNA response elements as allosteric modulators of steroid receptor function. Mol. Cell. Endocrinol. 2005;236:1–7. doi: 10.1016/j.mce.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Castet A, Herledan A, Bonnet S, Jalaguier S, Vanacker JM, Cavailles V. Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Mol. Endocrinol. 2006;20:1035–1047. doi: 10.1210/me.2005-0227. [DOI] [PubMed] [Google Scholar]
- 34.Darne CH, Morel L, Claessens F, Manin M, Fabre S, Veyssiere G, Rombauts W, Jean CL. Ubiquitous transcription factors NF1 and Sp1 are involved in the androgen activation of the mouse vas deferens protein promoter. Mol. Cell. Endocrinol. 1997;132:13–23. doi: 10.1016/s0303-7207(97)00116-0. [DOI] [PubMed] [Google Scholar]
- 35.Krishnan V, Wang X, Safe S. Estrogen receptor-Sp1 complexes mediate estrogen-induced cathepsin D gene expression in MCF-7 human breast cancer cells. J. Biol. Chem. 1994;269:15912–15917. [PubMed] [Google Scholar]
- 36.Porter W, Wang F, Wang W, Duan R, Safe S. Role of estrogen receptor/Sp1 complexes in estrogen-induced heat shock protein 27 gene expression. Mol. Endocrinol. 1996;10:1371–1378. doi: 10.1210/mend.10.11.8923463. [DOI] [PubMed] [Google Scholar]
- 37.Gottlicher M, Heck S, Herrlich P. Transcriptional cross-talk, the second mode of steroid hormone receptor action. J. Mol. Med. 1998;76:480–489. doi: 10.1007/s001090050242. [DOI] [PubMed] [Google Scholar]
- 38.Ponta H, Cato AC, Herrlich P. Interference of pathway specific transcription factors. Biochim. Biophys. Acta. 1992;1129:255–261. doi: 10.1016/0167-4781(92)90501-p. [DOI] [PubMed] [Google Scholar]
- 39.Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin HK, Hu YC, Yang L, Altuwaijri S, Chen YT, Kang HY, Chang C. Suppression versus induction of androgen receptor functions by the phosphatidylinositol 3-kinase/Akt pathway in prostate cancer LNCaP cells with different passage numbers. J. Biol. Chem. 2003;278:50902–50907. doi: 10.1074/jbc.M300676200. [DOI] [PubMed] [Google Scholar]
- 41.Cinar B, De Benedetti A, Freeman MR. Post-transcriptional regulation of the androgen receptor by Mammalian target of rapamycin. Cancer Res. 2005;65:2547–2553. doi: 10.1158/0008-5472.CAN-04-3411. [DOI] [PubMed] [Google Scholar]
- 42.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 43.Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]