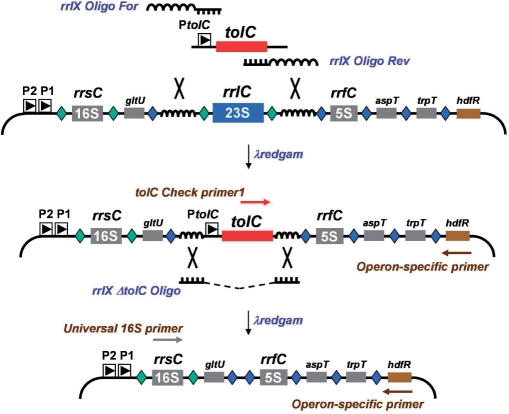
Figure 2.
Strategy for remodeling the Escherichia coli rRNA operons. A tolC cassette is shown amplified by rrlX oligos For and Rev (Rx-P50 and -P51, respectively) having universal homology arms that correspond to sequences flanking the 23S rRNA genes from all seven operons (Figure S3). In this example, removal of rrlC occurs following introduction of the tolC cassette and recombination. SDS-resistant colonies are screened by PCR using tolC Chk primer1 (Rx-P71) paired with an operon-specific primer (Px-P52 through P58). This screening can be multiplexed as primers were designed to amplify fragments of distinct sizes (Figure 3). Following confirmation of tolC insertion, the cassette is removed by introducing universal single-stranded oligos (RX-P59 or P60, depending on the direction of replication). After selecting for colicin E1 resistance, transformants are screened by plating on SDS and by PCR using a 16S rDNA primer with the operon specific primers mentioned before. Fragments of chromosomal DNA are depicted with bent lines. P2 and P1 refer to the promoters that drive expression of the rRNA operons and PtolC represents the P1 promoter for that gene. Neighboring genes are shown with shaded boxes: gltU, aspT and trpT encode tRNA; rrsC and rrfC encodes 16S and 5S rRNA, respectively. Green diamonds represent RNase III processing sites. Blue diamonds represent RNAse E processing sites for tRNA and 5S RNA. Primers used for PCR screening are shown with brown arrows. Curvy lines represent homologous DNA shared by the linear DNA substrate and the bacterial chromosome while large X's again denote λ red-mediated cross-over between regions of homologous DNA. Illustrations not drawn to scale.