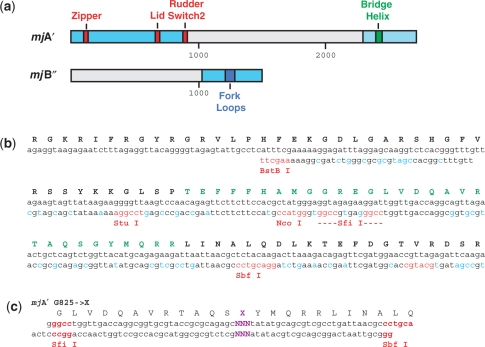
Figure 1.
Redesign of the bacterial expression vectors. (a) Schematic diagram of the redesigned coding regions of the mjA′ and mjB′′ bacterial expression vectors. The full-length open reading frames are shown with the codon-optimized sequences in light blue. The position of the sequences encoding key functional RNAP domains are shown in color (Zipper, Lid, Rudder, Switch-2 in red, Bridge Helix in dark green and Fork Loops in blue). The numbers refer to nucleotide positions. (b) Original and redesigned sequence encoding the Bridge Helix domain and surroundings (positions 2281-2550 in the mjA′ open reading frame). The original nucleotide sequence (as found in the M. jannaschii genome; 23) is shown on the top line with the redesigned sequence (starting at the BstBI site) beneath. The silently introduced restriction sites flanking various portions of the Bridge Helix-encoding sequence are in red, whereas other sequence alterations that improve codon usage in E. coli are shown in light blue. The amino acid sequence encoded is shown in single-letter code in black and the Bridge Helix primary sequence is highlighted in green. (c) Example of mutagenic oligonucleotides for creating targeted substitutions in Bridge Helix residues mjA′-G825. The two strands are designed to hybridize to each other to create a double-stranded ‘cassette’ bearing single-stranded extensions (shown in red) suitable for ligating into cloning vectors cleaved with suitable restriction enzymes. The cassettes shown have been used to replace the wild-type Bridge Helix sequence between the SfiI and SbfI sites (as shown in panel B) with a sequence containing a single randomized codon (shown in purple; 18).