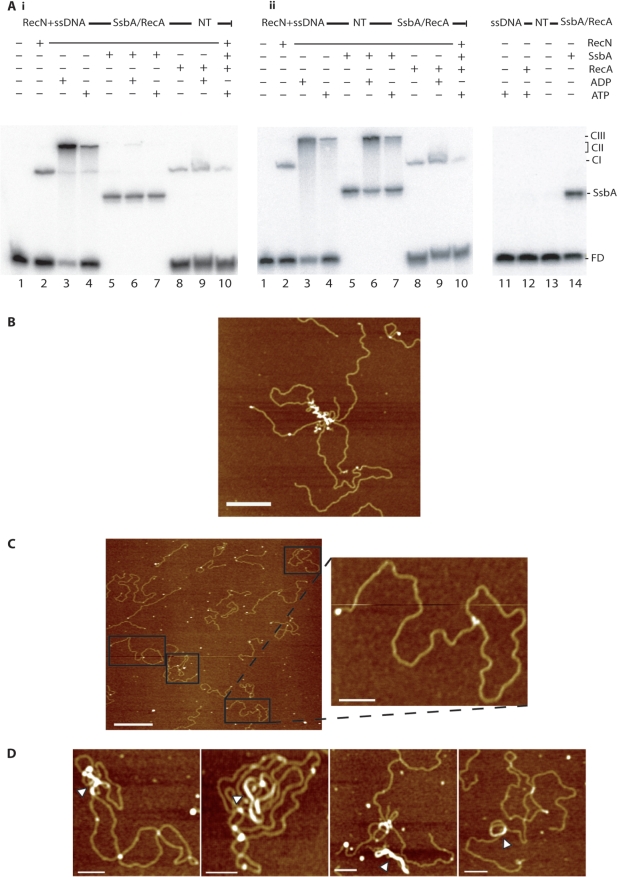
Figure 5.
RecA interaction with ssDNA–RecN–ATP–Mg2+ networks. (A) [γ-32P] ssDNA60 (60 nM in nt) and RecN (10 nM) were pre-incubated for 10 min at 37°C in buffer C. (Ai) SsbA (100 nM, lanes 5–7) or RecA (40 nM, lanes 8–10) protein was added to pre-formed RecN–ssDNA complexes and incubated for 10 min at 37°C, then 1 mM ADP (lanes 6 and 9) or 1 mM ATP (lanes 7 and 10) or no nucleotide cofactor (lanes 5 and 8) was added, and incubated for 10 min at 37°C. (Aii) 1 mM ADP (lanes 3, 6 and 9) or 1 mM ATP (lanes 4, 7 and 10) or no nucleotide cofactor (lanes 2, 5 and 8) was added to pre-formed RecN–ssDNA complexes, and then 100 nM SsbA (lanes 5–7) or 40 nM RecA (lanes 8–10) was added and incubated for 10 min at 37°C. The protein(s)–ssDNA–NT–Mg2+ complexes were fractionated and visualized. In the absence of RecN, [γ-32P] ssDNA60 (60 nM in nt) was incubated with RecA (40 nM, lanes 11 and 12) or SsbA (0.15 nM, lanes 13 and 14) in presence or absence of ATP. FD, free DNA; CI, complex I; CII, complex II; CIII, complex III; NT, presence of ADP or ATP; ±, denotes presence or absence of indicated factor. The SsbA–ssDNA complex (SsbA) is indicated. (B) RecA–DNA aggregates in the absence of RecN. Linear DNA (150 nM) with short ssDNA tails and 40 nM RecA were incubated for 10 min at 37°C, in the presence of 1 mM ATP at 37°C. Homologous circular plasmid DNA (150 nM) was added and incubated for another 10 min at 37°C, and deposited on freshly cut mica. Scale bar = 250 nm. (C) RecA-promoted DNA strand invasion in presence of RecN and ATP. Linear DNA (150 nM) with short ssDNA tails and 10 nM RecN were pre-incubated for 10 min at 37°C, then 1 mM ATP was added and incubated for 10 min at 37°C. Homologous circular plasmid DNA (150 nM) and RecA protein (40 nM) were added to the pre-formed ssDNA–RecN–ATP–Mg2+ complex, incubated for another 10 min at 37°C, and deposited on freshly cut mica. Scale bar = 500 nm (inset picture, bar = 100 nm). (D) RecA-promoted DNA strand invasion in presence of RecN and ATPγS. Linear DNA (150 nM) with short ssDNA tails and RecN (10 nM) were pre-incubated for 10 min at 37°C, then 0.1 mM ATPγS was added and incubated for 10 min at 37°C. Homologous circular plasmid DNA (150 nM) and RecA protein (40 nM) were added to the pre-formed ssDNA–RecN–ATPγS–Mg2+ complex, incubated for another 10 min at 37°C and deposited on freshly cut mica. White arrowheads show nucleoprotein filaments of RecA and DNA. Scale bar = 100 nm.