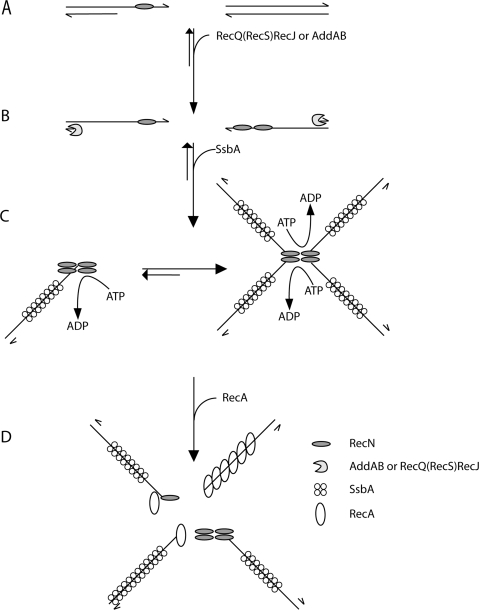
Figure 6.
Mode of action of RecN protein: a proposal. RecN binds ssDNA regions of broken DNA molecule (e.g. a single-strand nick in the lagging strand template) (A). In the presence of ATP–Mg2+ and prior DNA end-processing, RecN binds to the 3′-end of ssDNA tailed duplex DNA, because it binds with high efficiency to short ssDNA segments, whereas SsbA requires patches of ssDNA longer than 35-nt. The 5′ ends of the broken DNA molecules are processed either by RecJ in concerts with a RecQ-like protein (namely RecQ or RecS) or the AddAB helicase/nuclease, resulting in a duplex molecule with a 3′-terminated ssDNA tail (B). After end-processing, RecN by a protein–protein and protein–ssDNA interaction forms 1 to 2 RecN foci in vivo and large nucleoprotein networks (rosette-like structures) in vitro, and SsbA binds and protects the ssDNA (C). RecN and RecA interaction leads to disassembly of the rosette-like structures. In the absence of SsbA, RecN helps loading RecA onto naked ssDNA and RecA polymerizes onto ssDNA to form the active RecA nucleoprotein filament formation (D). In vivo RecN directly or indirectly interacts with RecO (and perhaps RecR), and the RecA-mediators might promote the disassembly of the SsbA protein and the loading of RecA onto ssDNA. The role of the RecA-mediators remains to be unravelled.