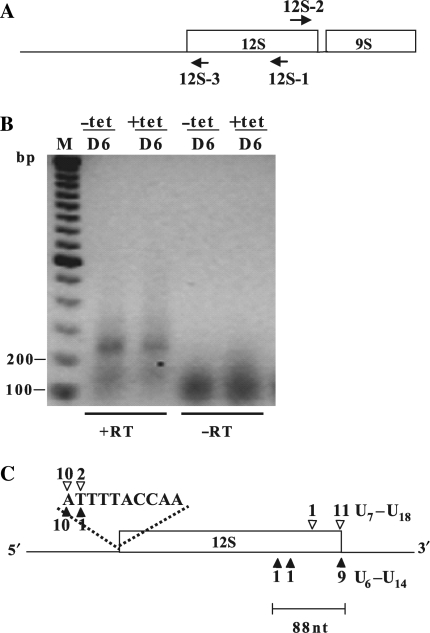
Figure 4.
TbDSS-1 is not required for correct 12S rRNA 3′ end processing. (A) Location of primers 12S-1, 12S-2 and 12S-3 used to map the 5′ and 3′ extremities of 12S rRNA in TbDSS-1 RNAi cells by cRT-PCR. Arrows indicate the orientations of the primers. (B) Negative image of an ethidium bromide-stained gel of cRT-PCR products amplified from 12S-1 primed cDNA using primer 12S-2 in combination with 12S-3. RNA was isolated from uninduced cells (−tet) and cells induced with tetracycline for 6 days (+tet). M, DNA marker consisting of bands with 100 bp increments. The positions of the 100 and 200 bp makers are indicated on the left. Other labels as in Figure 2. (C) Schematic of experimentally determined 5′ and 3′ extremities. Solid arrowheads indicate RNAs from induced cells; open arrowheads indicate RNAs from uninduced cells. The number of clones obtained at a given site is indicated above or below each arrowhead. The lengths of the oligo(U) tails of those clones with a mature 3′ extremity are indicated.