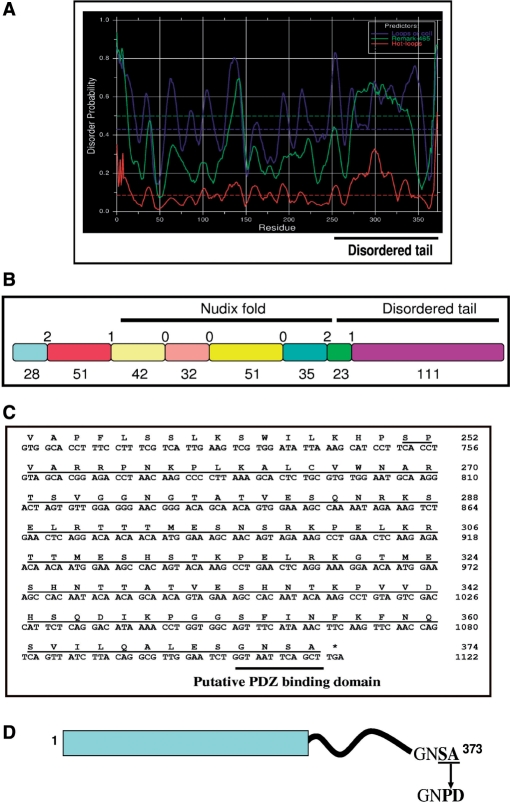
Figure 2.
Bioinformatics analysis predicts region of structural disorder and existence of a putative PDZ binding domain in the AtDcp2 sequence. (A) Prediction of disordered regions using the DisEMBL server—in the graphical output from DisEMBL, the disorder prediction is calculated using the three criteria: (i) secondary structural types of loops/coils (blue), (ii) hot loops which constitute a subset of loops comprising inherent high mobility (red) and (iii) missing coordinates in X-ray structures (green). (B) Schematic representation of the protein fragments encoded by each exon of the At5g13570 gene coding for AtDcp2 mRNA decapping enzyme—labels indicate the size of proteins fragments encoded by each exon (below) and the intron phases (above) of the At5g13570 gene. The positions of the Nudix fold and the disordered tail are shown. (C) The sequence corresponding to the disordered region of the AtDcp2 protein (underlined) with emphasis on the putative PDZ domain-binding motif. (D) The putative PDZ domain-binding motif in the wild-type AtDcp2 protein and mutations introduced to generate the recombinant GST-AtDcp2-PD and GST-AtDcp2-PD-His10 proteins.