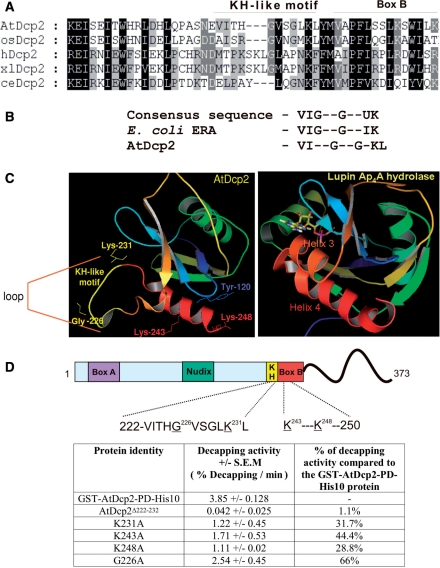
Figure 7.
Effects of mutation of residues within the putative RNA-binding KH-like domain and Box B domain on catalytic activity of AtDcp2. (A) Sequence alignment of Arabidopsis thaliana (AtDcp2) to reveal the putative KH-like domain. Oryza sativa (osDcp2), human (hDcp2), Xenopus laevis (xlDcp2) and Caenorhabditis elegans (ceDcp2) Dcp2 decapping enzymes were used for the alignments. The position of the KH-like motif and Box B within the AtDcp2 protein is shown in the sequence alignment. (B) The consensus sequence of the conserved 10 residue region found in KH domains (42,43), the sequence corresponding to the consensus sequence found in the KH domain in the RNA-binding site of ERA protein from E. coli (42) and the sequence of a KH-like motif in the AtDcp2 decapping enzyme. (C) Comparison of the structure of the Ap4A hydrolase from L. angustifolius (PDB ID–1JKN) with the homology model of the Nudix fold of the AtDcp2 protein (residues 98–250) modelled after the structure of the S.pombe Dcp2p enzyme (PDB ID – 2A6T). Box B helix in the AtDcp2 homology model and the analogous helix 4 in the lupin enzyme structure are marked in red. The 11-residue region forming the near-consensus of a KH motif, found within the loop region preceding Box B, is coloured in yellow. This region was deleted in mutant AtDcp2Δ222–232. The side chains of three lysines and a single glycine, which were mutated individually to alanines, are also shown. (D) Decapping activities of the GST-AtDcp2-PD-His10 protein and the mutant proteins carrying mutations in the putative RNA-binding site. All decapping efficiencies were obtained from decapping assays carried out using 8 μg of recombinant enzyme from three separate expressions. SEM, standard error of mean.