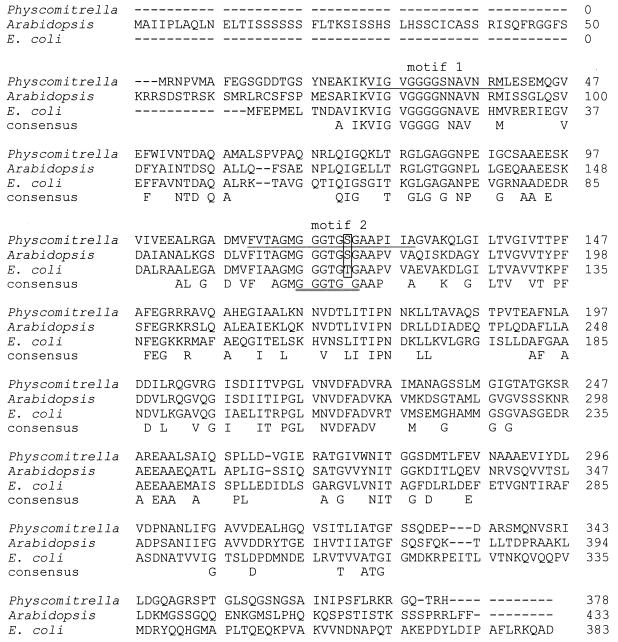
Figure 1.
Alignment of amino acid sequences of two eukaryotic and one prokaryotic FtsZ proteins in the one-letter-code. Aligned are PpFtsZ from the moss Physcomitrella patens (GenBank AJ001586), cpFtsZ from the dicotyledon Arabidopsis thaliana (SwissProt Q42545) and FtsZ from the enterobacterium Escherichia coli (SwissProt P06138). Where applicable a consensus is given below the alignment. Two PROSITE motifs, FTSZ 1 and FTSZ 2, are marked by underlining the Physcomitrella sequence once. A third PROSITE motif, the tubulin signature motif, is marked by underlining the consensus sequence twice. An amino acid position is boxed within this motif, where serines in the eukaryotic proteins match perfectly to the tubulin signature motif while a threonine in the prokaryotic protein deviates from this consensus motif. Note the different lengths of the N-terminal extensions of the two eukaryotic FtsZ proteins as compared with the bacterial sequence.