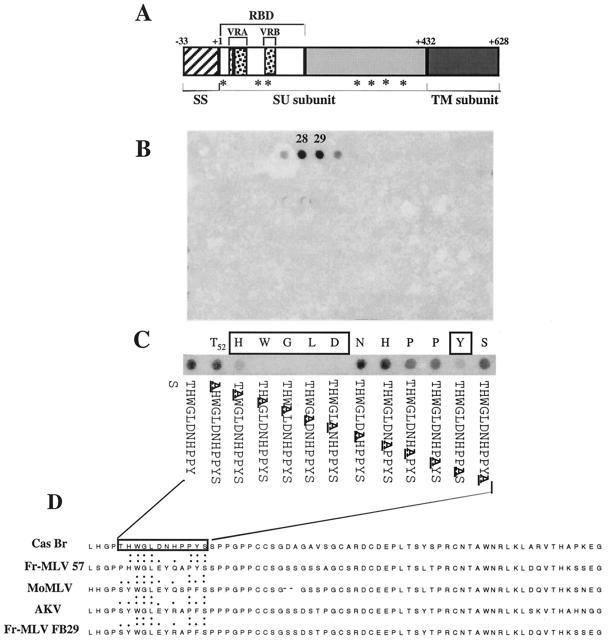
FIG. 2.
Characterization of the 667 epitope. (A) Structure of the CasBrE Env precursor. CasBrE is synthetized as a gp85 precursor which is proteolytically processed into a gp70 surface protein (SU) and a p15(E) transmembrane protein (TM). RBD, receptor binding domain; SS, signal sequence. VRA and VRB are the two most variable domains among the different Envs. Numbers indicate amino acid positions. Asterisks indicate CasBrE Env putative glycosylation sites. The black box represents the location of the epitope recognized by MAb 667. (B) Spot analysis of CasBrE Env. A total of 217 overlapping 15-mer oligopeptides were synthetized on a cellulose membrane and probed with a 667 MAb-containing hybridoma cell culture supernatant as described in Materials and Methods. (C) Alanine scanning of the 667 epitope. A series of twelve 15-mer oligopeptides, each containing one amino acid change into alanine at a different position, was synthetized on a cellulose membrane and probed with MAb 667. (D) Amino acid comparison of various ecotropic MLV Envs. The VRA regions from five ecotropic MLV Envs are compared. The 667 epitope is enclosed in a box. Single and double points indicate amino acids homologous and identical to CasBrE Env amino acids, respectively.