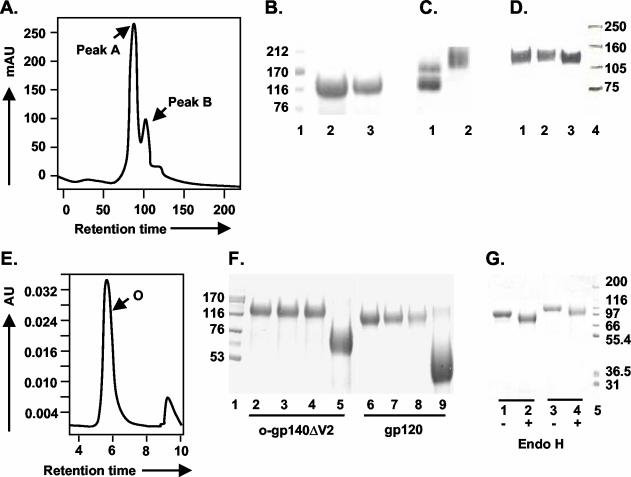
FIG. 5.
Purification and glycosylation linkage analysis of o-gp140SF162ΔV2. (A) gp140SF162ΔV2 obtained after the CHAP column was further fractionated on a precalibrated Superdex-200 sizing column to separate oligomers from the dimers and/or monomers of gp140. Peaks corresponding to oligomer (peak A) and dimer or monomer (peak B) are indicated. (B and C) Polyacrylamide gel analysis of the sizing fractions in reducing and denaturing conditions (B) (lanes: 1, molecular weight standards; 2, o-gp140SF162ΔV2; 3, gp140SF162ΔV2 dimer/monomer) and native conditions (C) (lanes: 1, dimer and monomer; 2, oligomer). (D) Immunodetection of o-gp140 using a MAb (20-2-C8.5F3) directed against the C4 domain of gp120 SF2 (lanes: 1, o-gp140SF162ΔV2;2, gp140SF162ΔV2 monomer; 3, gp120SF2). (E) Size exclusion-HPLC profile of the purified o-gp140SF162ΔV2. (F) Carbohydrate linkage analysis of purified o-gp140SF162ΔV2. Lanes: 3 and 7, o-gp140SF162ΔV2 and gp120SF162 digested with NANase; 4 and 8, O-glycosidase; 5 and 9, PNGF; 2 and 6, o-gp140SF162ΔV2 and gp120SF162 without any enzyme, used as controls; 1, molecular weight standards. (G) Endo-H digestion of gp120SF162 (lanes 1 and 2) and o-gp140SF162ΔV2 (lanes 3 and 4). + and −, presence or absence of Endo-H.