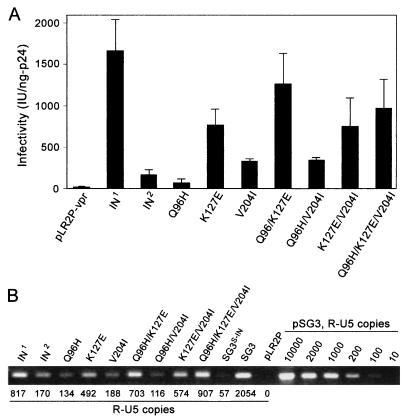
FIG. 5.
Analysis of mutations selected in the IN2 coding region by trans-complementation; 4 μg of pSG3S-IN proviral DNA was cotransfected into 293T cells with 6 μg of pLR2P-vpr, pLR2P-vprIN1, or pLR2P-vprIN2 (controls) or with 6 μg of the pLR2P-vprIN expression plasmids containing mutations in the IN coding region, as depicted. Forty-eight hours after transfection, the culture supernatants were collected, clarified by low-speed centrifugation, filtered through 0.45-μm sterile filters, and analyzed for virus concentration by HIV-1 p24 antigen ELISA. (A) Analysis of virus infectivity. Fivefold serial dilutions of each virus were prepared and analyzed for infectivity with TZM-bl reporter cells. Infectivity was standardized to 1 ng of p24 antigen. (B) Analysis of viral DNA synthesis; 500 ng (p24 antigen equivalents) of each virus was analyzed as described for Fig. 2B. A standard curve was created with standards ranging from 10 to 10,000 copies of pSG3 DNA.