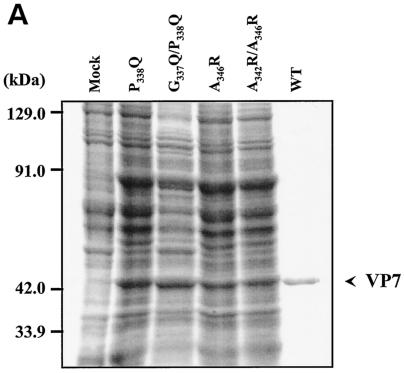
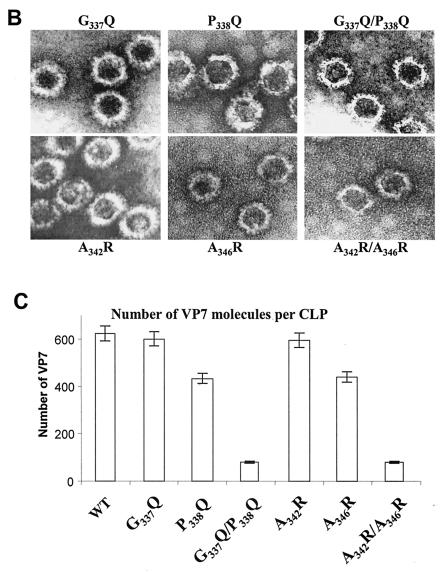
FIG. 5.
Location and effects of mutations at conserved VP7 residues within alpha helix 9. (A) Analysis of infected cell lysates by four (P338Q, G337Q/P338Q, A346R, and A342R/A346R) of the six mutant viruses by SDS-10% PAGE, followed by Coomassie blue staining. Analysis of coexpression of VP3 with wild-type VP7 (WT) or with mutants (G337Q, P338Q, A342R, A346R, G337Q/P338Q, and A342R/A346R) was done as indicated. (B) EM analysis of CLPs formed by native VP3 protein and native VP7 or different mutant VP7 as indicated. (C) The numbers of VP7 molecules per CLP under high-salt conditions and calculated as described for Fig. 2 are given. The data shown are representatives of three experiments, the value of which varied by no more than 10%.