Figure 1.
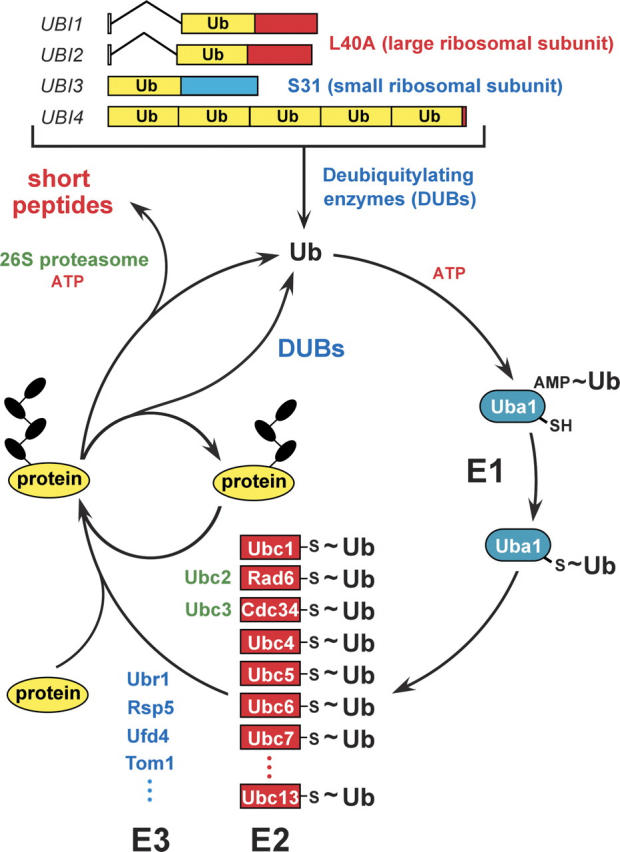
The ubiquitin system of the yeast S. cerevisiae (Varshavsky 1997, 2005). The fundamental design of this system is conserved among eukaryotes. The yeast ubiquitin genes UBI1–UBI4, two of which contain introns, encode fusions of ubiquitin either to itself or to one of two ribosomal proteins. These fusions are cleaved by deubiquitylating enzymes (DUBs), yielding mature ubiquitin. Thioester bonds between ubiquitin and the active-site Cys residues of ubiquitin-specific enzymes are denoted by the “~” sign. The conjugation of ubiquitin to other proteins involves a preliminary ATP-dependent step, in which the last (Gly-76) residue of ubiquitin is joined, via a thioester bond, to a Cys residue of the ubiquitin-activating (E1) enzyme, encoded by UBA1. The activated ubiquitin is transferred to a Cys residue in one of several ubiquitin-conjugating (E2) enzymes, encoded by the UBC-family genes, and from there to a Lys residue of an ultimate acceptor protein. This last step, and the formation of a substrate-linked polyubiquitin chain (black ovals) require participation of another component, called E3, whose mechanistic functions include the recognition of a substrate’s degradation signal (degron). The names of some of the currently known yeast E3s are indicated as well. The term “ubiquitin ligase” denotes either an E2–E3 holoenzyme or its E3 component. A targeted, ubiquitylated protein is processively degraded to short peptides by the ATP-dependent 26S proteasome. (For reviews, see Varshavsky 1997; Hershko et al. 2000; Pickart 2004.)
