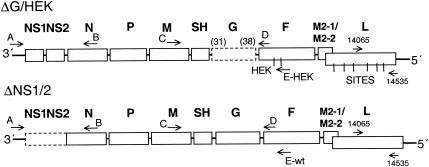
FIG. 1.
Gene maps of the two rRSV mutants, ΔG/HEK and ΔNS1/2, used for coinfection of HEp-2 cells. Genes are identified on top by the proteins which they encode: NS1 and NS2, nonstructural proteins 1 and 2; N, nucleocapsid protein; P, phosphoprotein; M, matrix protein; SH, small hydrophobic protein; G, attachment protein; F, fusion protein; M2-1, transcription antitermination factor; M2-2, RNA synthesis factor (M2-1 and M2-2 are encoded by two overlapping open reading frames in the same M2 mRNA); L, large polymerase protein. HEK and SITES are point mutations in the F and L genes, respectively, that are unique to the ΔG/HEK mutant (see Materials and Methods). Dashed boxes indicate the gene deletions of each rRSV mutant. The SH-F junction in ΔG/HEK contained the upstream 31 nt of the original SH-G junction fused to the downstream 38 nt of the original G-F junction (numbers in parentheses). In ΔNS1/2, the ATG of the N open reading frame was fused to the upstream nontranslated region of the NS1 gene. The placement of primers (A, B, C, D, E-HEK, E-wt, 14065, and 14535), used to differentiate the mutant constructs by RT-PCR (Table 1 and Materials and Methods), is indicated by arrows.